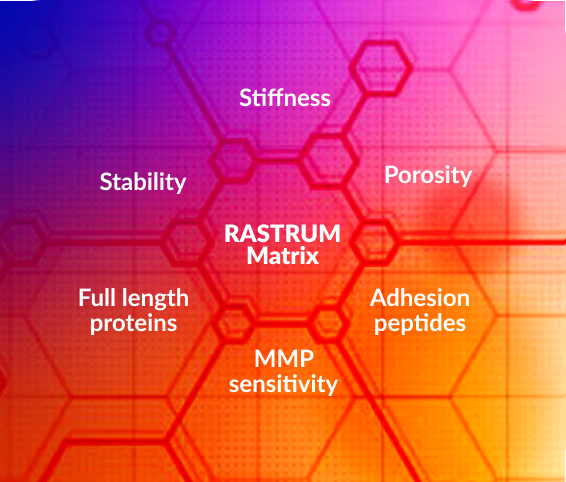
Advancing cancer research with 3D cell models
Understanding cancer starts with the right model. Traditional 2D cultures and animal models fail to capture the full complexities of human tumors, leading to poor clinical translation. The RASTRUM™ 3D cell culture platform overcomes these limitations—enabling researchers to build reproducible, scalable, and biologically-relevant tumor microenvironments that drive breakthrough discoveries in oncology.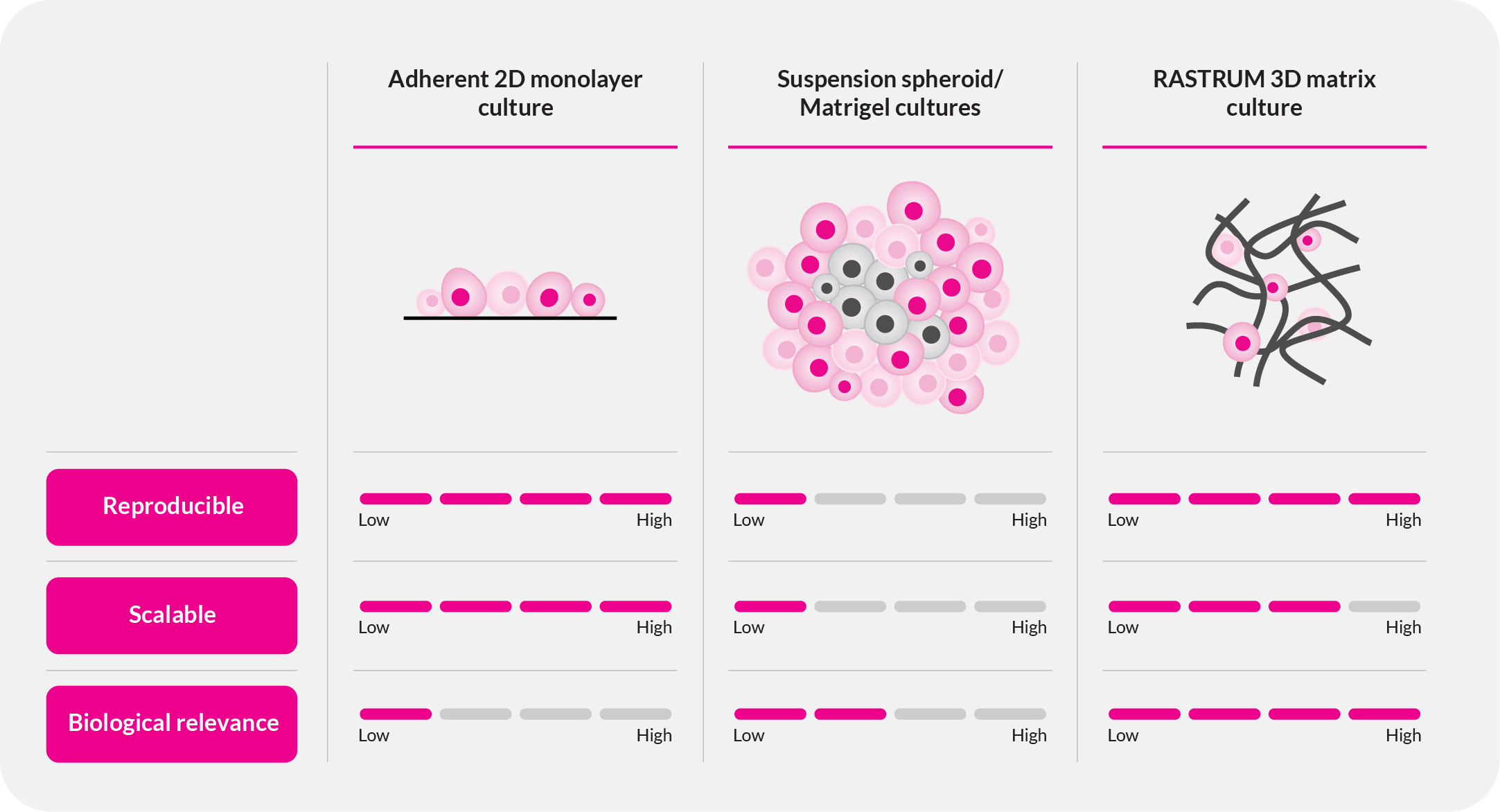
The challenge: Bridging the gap in cancer research
Cancer is dynamic, complex, and highly heterogeneous, yet many research models fail to reflect the true biology of tumors.
- 2D cultures lack the diversity of critical cell-cell and cell-matrix interactions, limiting their relevance.
- Animal models are expensive, time-consuming, and often fail to predict human responses.
- Traditional approaches suffer from variability, leading to inconsistent and unreliable results.
RASTRUM changes this paradigm. By enabling scalable, reproducible, and phenotypically relevant 3D cell models, we empower cancer researchers to study tumor biology with unprecedented accuracy and translational relevance.
RASTRUM: A revolution in 3D cancer research
RASTRUM enables the rapid creation of complex, reproducible 3D tumor models that mimic the biology present in the tumor microenvironment—providing researchers with a more predictive system for studying cancer biology and treatment response.
Why RASTRUM for Cancer Research?
- Phenotypically relevant tumor models: Capture tumor heterogeneity and microenvironment interactions.
- Advanced co-cultures: Create heterogeneous tumor models with controlled spatial separation or integrated cell populations to study cell-cell interactions in cancer progression.
- Scalable and reproducible: High-throughput, standardized tumor architectures for consistent results.
- Seamless integration with drug screening workflows: Optimized for preclinical pipelines.
RASTRUM empowers researchers to accelerate discoveries in tumor biology, drug development, and immuno-oncology—bridging the gap between in vitro studies and clinical translation.
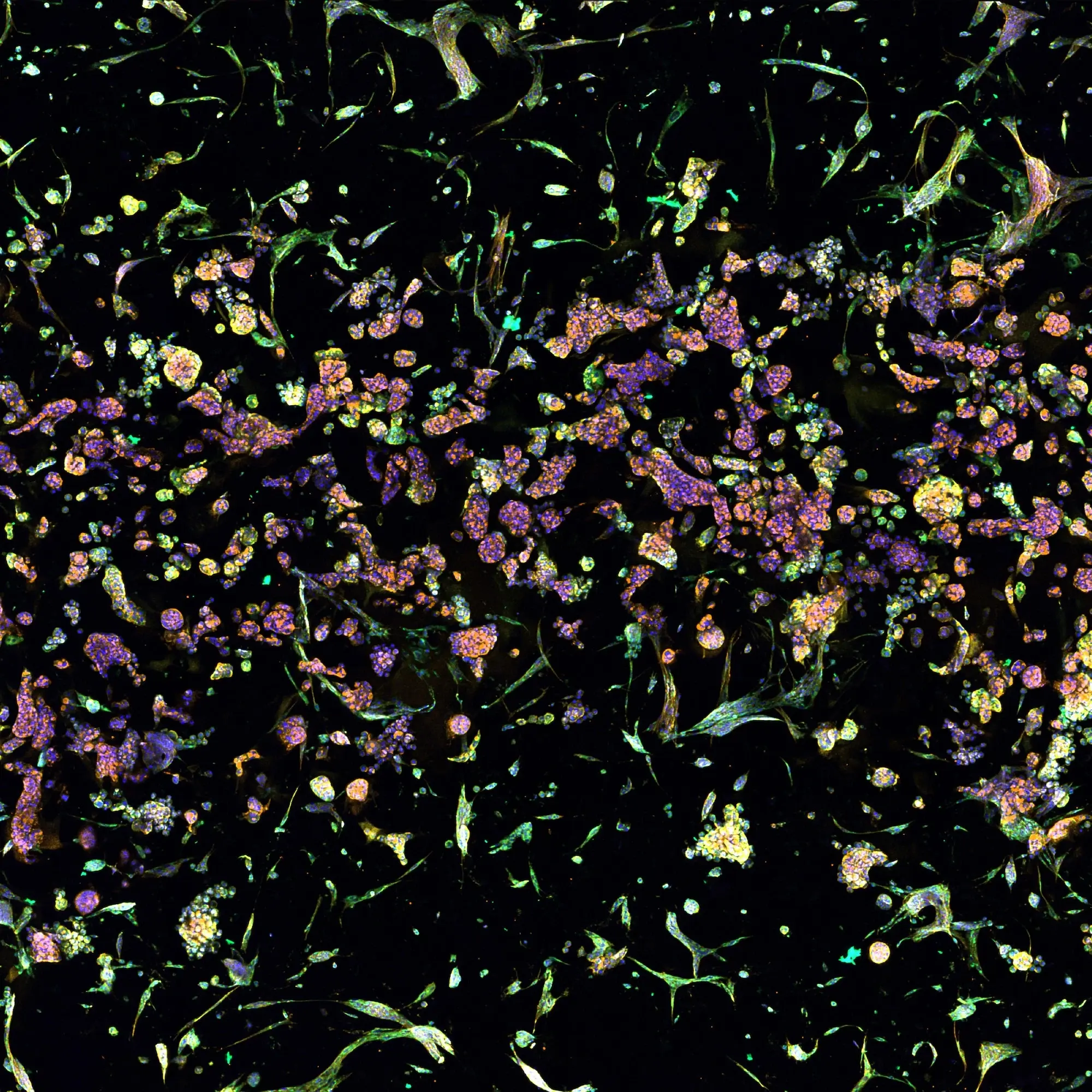
The figure depicts a lung cancer tri-culture model made with RASTRUM Allegro. Primary normal human lung fibroblasts (NHLFs) were printed with primary Human Umbilical Vein Endothelial Cells (HUVECs) and A549 lung adenocarcinoma cells using RASTRUM’s Imaging Model architecture. Cells were cultured for 7 days and stained using a PhenoVue Cell Painting Kit (Revvity). Cell nuclei (cyan) were labeled with Hoescht, filamentous actin (yellow) with phalloidin, and membrane glycoproteins (magenta) with concanavalin A.
3D cell model applications in cancer research
Tumor microenvironment modeling with 3D cancer models
Creating phenotypically relevant tumor models is essential for understanding cancer progression, drug resistance, and development of new therapeutic approaches. RASTRUM enables researchers to engineer complex tumor-stroma interactions that are critical for studying the biology of solid tumors.
- Recreate native tumor-stroma interactions: Model the interplay between key cell types such as cancer cells, fibroblasts, immune cells, and extracellular matrix components.
- Investigate fibrosis, hypoxia, and immune infiltration: Generate tumor models with varied stromal stiffnesses, and immune cell recruitment.
- Study cancer progression and treatment resistance: Analyze how changes in the microenvironment drive tumor evolution, therapy resistance, and metastatic potential.
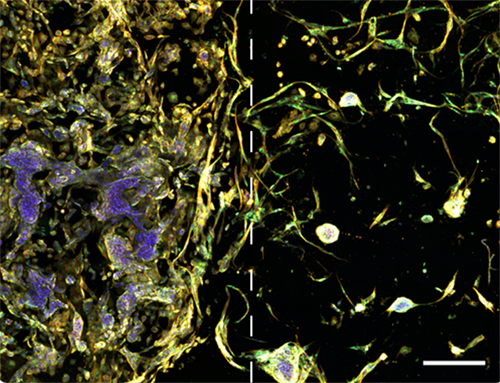
Image illustrates tumor microenvironment with spatial separation in RASTRUM’s Dual Matrix Model architecture. Lung cancer (A549) cells (left) and normal human lung fibroblast (right) were printed in a Dual Matrix Model architecture on RASTRUM and imaged after seven days in vitro. Dotted lines indicate the boundaries between the matrices. Cells were labelled using the PhenoVue Cell Painting Kit: nuclei (blue, Hoechst), endoplasmic reticulum (green, concanavalin A), and actin (yellow , phalloidin).
Immuno-oncology research with 3D tumor models
Understanding the interplay between tumors and the immune system is critical for developing next-generation cancer immunotherapies. RASTRUM enables researchers to model complex tumor-immune interactions, providing a phenotypically relevant platform to evaluate immune-based treatments.
- Model tumor-immune interactions: Incorporate T cells, macrophages, dendritic cells, and NK cells into 3D tumor models to study immune infiltration, activation, and suppression.
- Evaluate CAR-T and checkpoint inhibitor therapies: Assess the efficacy of immune checkpoint blockade, adoptive T cell therapy, and biotherapeutics in a physiologically relevant tumor environment.
- Predict immune evasion and resistance mechanisms: Investigate how tumors suppress immune responses, modulate cytokine signaling, and develop resistance to immunotherapies.
- Study the tumor microenvironment’s impact on therapy: Examine how stromal cell cross-talk, stromal barriers, and immune cell exhaustion affect treatment outcomes.
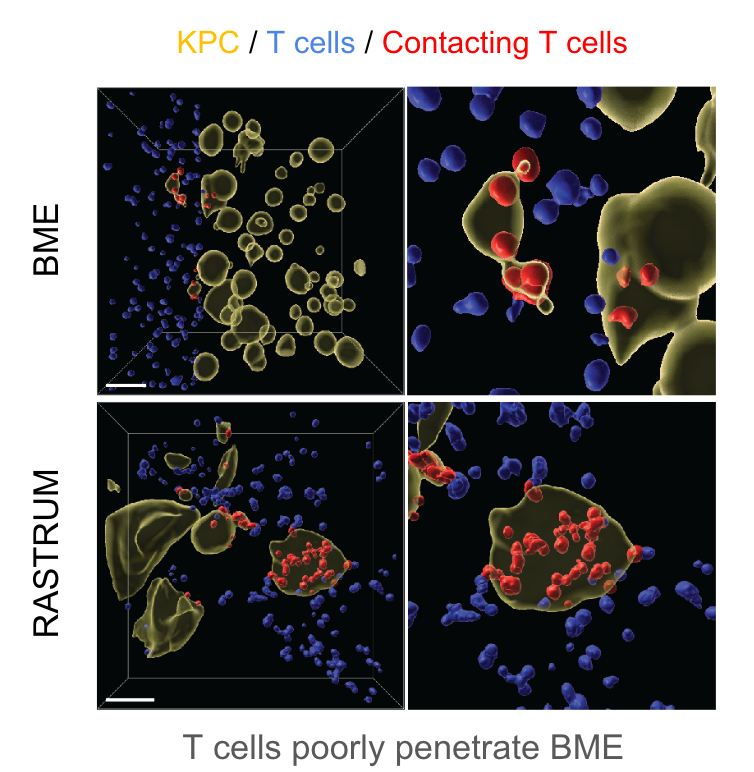
The figure depicts pancreatic cancer cells cultured in 4.8kPa matrices for 3 days before 12,000 activated cytotoxic T lymphocytes (CTLs) were added to the media in each well. Imaging and Imaris analysis shows CTL infiltration 2 days post-addition and allows quantification of interactions (red) between cancer cells (yellow) and T cells (blue) in RASTRUM matrices and basement membrane extract (BME).
.png)
Drug discovery and screening using 3D cancer models
Successful drug development depends on models that accurately predict clinical outcomes. Traditional in vitro and in vivo approaches often fail to capture the complexity of human tumors, leading to high attrition rates in drug pipelines. RASTRUM enables scalable, phenotypically relevant 3D tumor models that integrate seamlessly into drug discovery workflows, improving efficiency and success rates.
- Accelerate lead identification and optimization: Generate reproducible 3D tumor models to screen drug candidates earlier in the pipeline.
- Improve translational relevance: Model tumor heterogeneity, cell-cell interactions, and microenvironment effects to better predict clinical outcomes.
- Enhance high-throughput screening: Conduct large-scale drug testing with standardized, scalable tumor models, reducing variability and increasing confidence in early-stage results.
- Reduce false positives and negatives: Improve decision-making by minimizing reliance on poorly predictive 2D and animal models.
The figure shows MCF-7 breast cancer cells cultured in 1.1kPa matrices for 6 days before addition of cisplatin to wells at 1μM (left) and 100μM (right). Live/dead staining and imaging was performed 24 hours post-addition of cisplatin, with live cells shown in green (Calcein-AM) and dead cells in red (EthD-III).
Precision medicine research with 3D tumor models
Advancing precision medicine starts with understanding how individual tumors respond to treatment. Traditional models often fail to capture patient-specific tumor biology, limiting their predictive value in drug development. RASTRUM enables researchers to generate phenotypically relevant 3D tumor models to study tumor heterogeneity, identify biomarkers, and improve preclinical drug evaluation.
- Develop patient-relevant tumor models: Use primary patient-derived cells to study tumor-specific responses in a controlled 3D microenvironment.
- Identify biomarkers for targeted therapies: Investigate molecular and phenotypic markers that correlate with drug sensitivity and resistance.
- Improve preclinical drug evaluation: Generate reproducible 3D models that better reflect in vivo conditions, reducing reliance on less predictive systems.
- Support research into therapy resistance and tumor evolution: Study how tumors adapt over time and explore mechanisms of drug resistance in a scalable and reproducible platform.
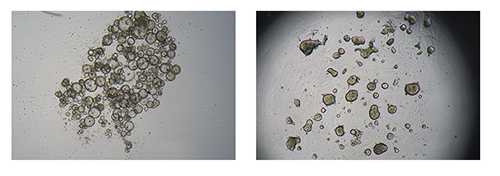
The figure depicts bright-field images of patient-derived colorectal cancer cells grown as tumoroids in suspension cultures at day 6 (left) or in Px02 RASTRUM matrices at day 7 (right).
Metastasis and invasion studies with 3D cell models
Metastasis is the leading cause of cancer-related mortality, yet traditional models struggle to accurately capture the cellular and microenvironmental dynamics of tumor invasion and dissemination. RASTRUM enables researchers to create phenotypically relevant 3D models that mimic metastatic progression, providing a powerful tool for studying invasion mechanisms and testing potential therapeutic strategies.
- Recreate cancer cell invasion and metastatic spread: Model tumor cell migration and invasion within a controlled 3D environment.
- Study epithelial-mesenchymal transition: Investigate the molecular and phenotypic changes driving tumor cell plasticity, migration, and drug resistance.
- Examine tumor-stroma interactions in metastasis: Explore how fibroblasts, immune cells, and extracellular matrix composition influence metastatic potential.
- Evaluate potential anti-metastatic strategies: Test compounds that disrupt invasion pathways, prevent tumor dissemination, and target metastatic niches in a reproducible, high-throughput system.
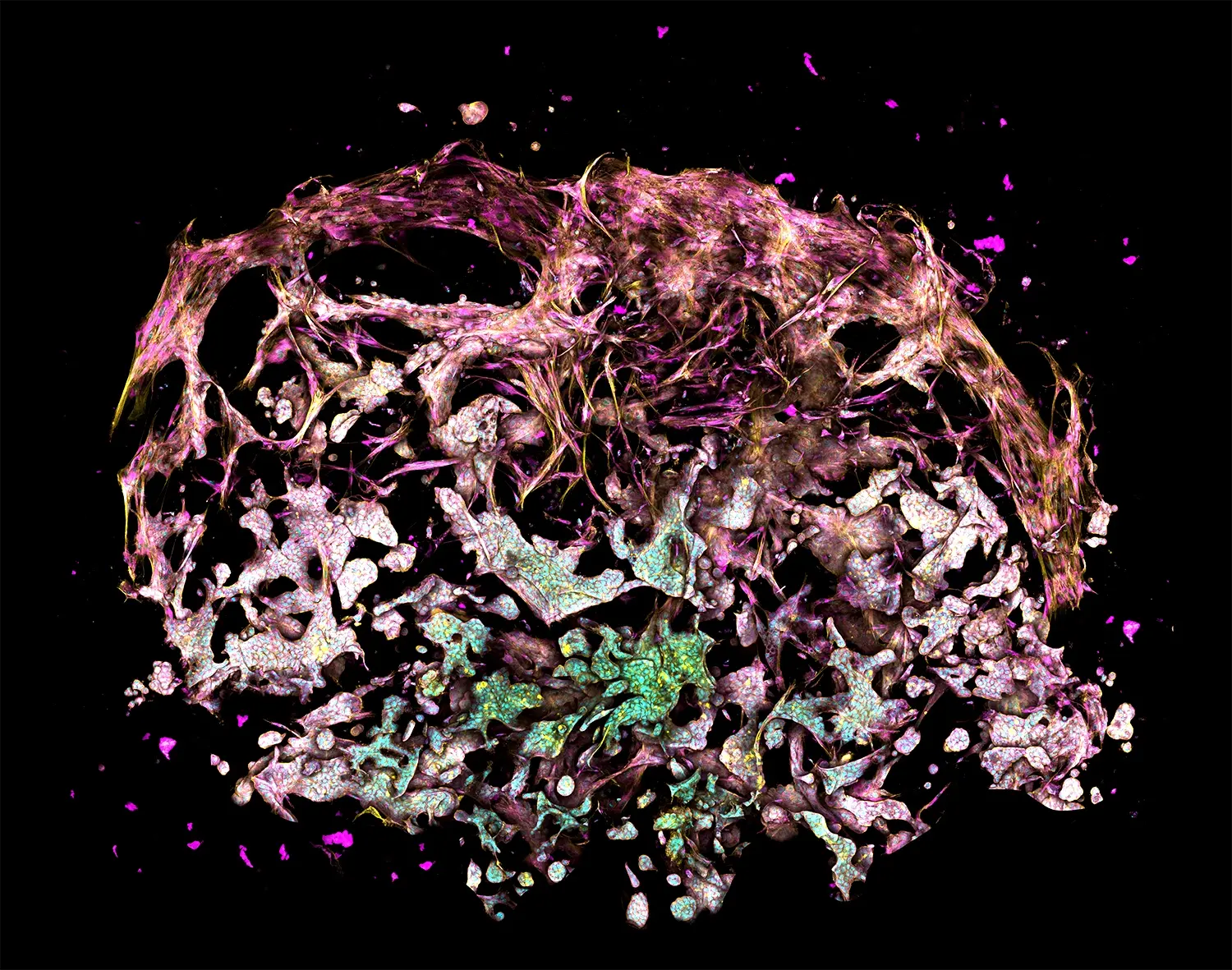
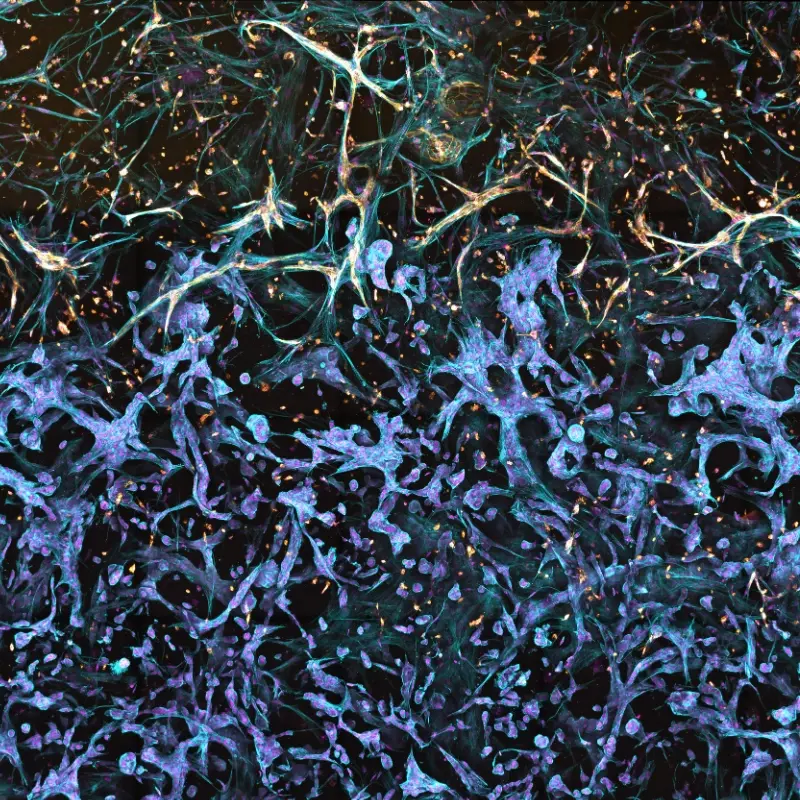
The figure depicts a model of the tumor-stroma interface in a Triple Matrix Model made with RASTRUM. Fibroblasts (primary human lung fibroblasts) and endothelial cells (primary HUVECs) were grown next to lung adenocarcinoma cancer cells (A549) and fibroblasts (HUVECs) were cultured for 10 days. Endothelial cells were stained with CD31 (yellow) and fibroblasts and endothelial cells were stained with Phalloidin (blue).
FAQs
3D cancer models are laboratory-engineered tumor models that mimic the complexity of human tumors. Unlike traditional 2D cultures, they capture cell-cell interactions, extracellular matrix dynamics, and tumor heterogeneity, providing more physiologically relevant insights for cancer research and drug discovery.
RASTRUM enables the creation of scalable and reproducible 3D tumor models for use in disease research and drug discovery. The approach enables the generation of simple or complex co-cultures with multiple cell types, including tumor, stromal, and immune cells, the ability to mimic tumor heterogeneity and microenvironment dynamics, and standardization of high throughput production of 3D cancer models for consistent and reproducible results. By eliminating the variability of manual 3D culture methods, RASTRUM provides a more reliable and scalable solution for cancer research.
RASTRUM overcomes key limitations of 2D cultures, spheroid models, and animal studies by providing:
- More predictive results: Better replicates tumor biology, improving translational relevance.
- Scalability and reproducibility: Speeds up 3D model generation, reducing variability within and between experiments.
- Advanced co-culture capabilities: Supports tumor-stroma-immune interactions in defined spatial architectures.
- Drug discovery screening: Enables high-throughput screening with standardized tumor models.
Yes. RASTRUM is designed for high-throughput 3D cell model generation, allowing researchers to rapidly produce thousands of 3D tumor models in parallel. These models provide a phenotypically relevant tumor environment for screening drug candidates. RASTRUM 3D cell models seamlessly integrate with automated imaging and analysis pipelines, accelerating the discovery process and enabling large-scale studies. By combining scalability and reproducibility, RASTRUM supports efficient, high-throughput drug discovery efforts while ensuring data consistency across experiments.