
Ready-to-use cell model architectures optimized for your applications
Empower your research with a range of 3D cell model architectures tailored to your needs. Seamlessly integrate these flexible architectures into various well-plate formats and applications, including imaging, screening, and cell extraction.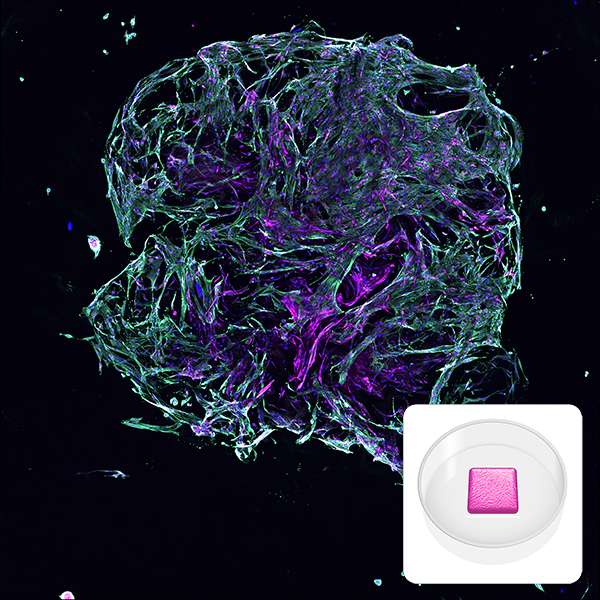
Imaging Model
A small single-matrix architecture optimized for fast imaging and minimal cell usage. This model ensures clear imaging and compatibility with standard techniques, enabling seamless integration into your workflows. Compatible with 96-well plates on RASTRUM™ and RASTRUM™ Allegro.
Applications
- Brightfield and immunofluorescence imaging
- Drug screening
- Biochemical assays
Features
- Thin architecture for rapid, high-content imaging
- Compact design minimizes the number of cells required while maintaining reproducibility
Image depicts a lung fibroblast model made with RASTRUM. Primary normal human lung fibroblasts (NHLFs) were printed using RASTRUM’s Imaging Model architecture. Cells were cultured for 7 days and immunostaining was performed to visualize collagen I (magenta), alpha smooth muscle actin (green) and cell nuclei (blue). Inset: Imaging Model architecture illustration.
Large Plug Model
A single-matrix architecture tailored for expansion and bulk downstream analyses, delivering the cellular material you need for comprehensive studies and high-throughput applications. Compatible with 96-well plates on RASTRUM and RASTRUM Allegro.
Applications
- DNA, RNA, and protein analysis
- Flow cytometry
- Omics analyses
Features
- Larger matrix architecture for increased cellular material
- Optimized for workflows requiring bulk analysis and high data yield
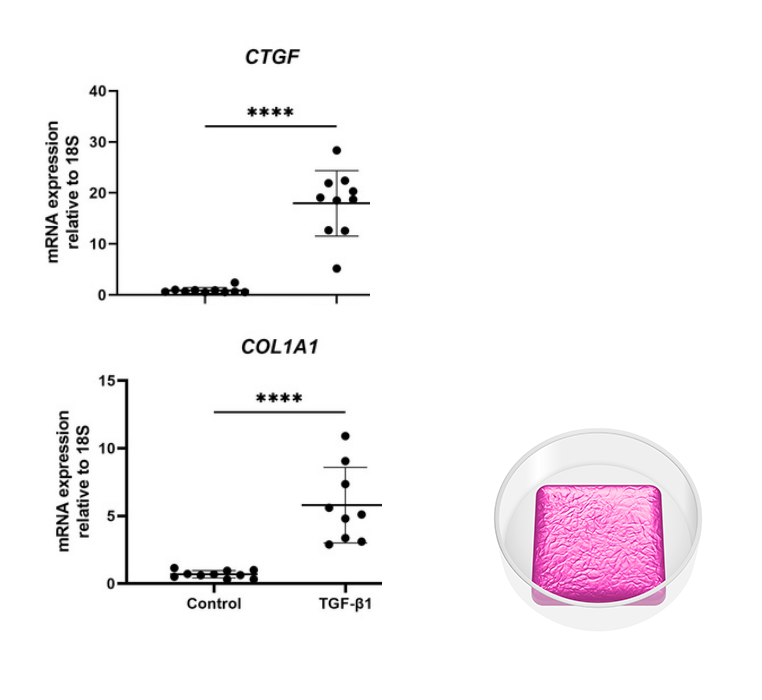
Image depicts analysis of a liver co-culture created with RASTRUM. Immortalized hepatocyte and stellate cells were printed in a Large Plug Model and cultured for 5 days before treatment with TGF-β1. On Day 7 cells were retrieved from the matrix and RNA isolated. Gene expression of profibriotic markers for Collagen-type 1 (COL1A1) and connective tissue growth factor (CTGF) were shown to be significantly increased following TGF-ꞵ1 treatment. Inset: Large Plug Model architecture illustration.
Screening Model
A single-matrix architecture optimized for high-throughput workflows, enabling efficient data collection and scalable experimentation. Compatible with 384-well plates on RASTRUM Allegro.
Applications
- Drug screening
- Cytotoxicity assays
- Mechanistic studies
Features
- Optimized for assay flexibility and adaptability
- Consistent and reproducible results across screening workflows
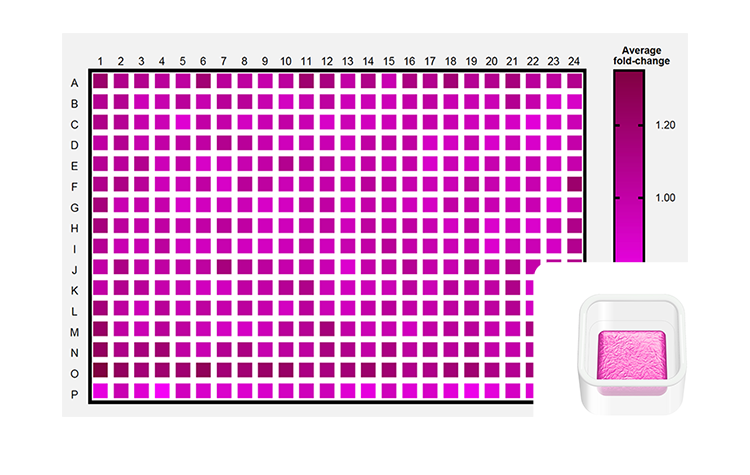
Image depicts consistent MCF-7 drug sensitivities at scale. MCF-7 cells printed in the Screening Model architecture in wells of twelve 384-well plates across two independent PrintRuns. Low post-print and drug treatment CVs were observed. Inset: Screening Model architecture illustration.
High-Throughput Model
A single-matrix architecture optimized for high-throughput workflows in 384-well plates, enabling efficient data collection and scalable experimentation. Compatible with 384-well plates on RASTRUM.
Applications
- Drug screening
- High-content imaging
- Biochemical assays
Features
- Consistent and reproducible 3D models in 384-well plate format
- Streamlined for large-scale assays and automated workflows
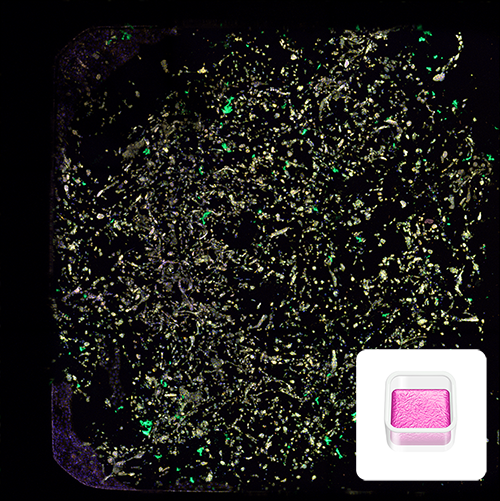
Image depicts a 3D culture of a metastatic mammary adenocarcinoma. Matrix cultures with MDA-MB-231 cells were generated with RASTRUM using the High-Throughput Model architecture, and grown for 7 days in 384 well plates. Morphology staining was applied for high content phenotypic imaging using the PhenoVue Cell Painting Kit: nuclei (blue, Hoechst), endoplasmic reticulum (green, concanavalin A), and actin (yellow , phalloidin). Inset: High-Throughput Model architecture illustration.
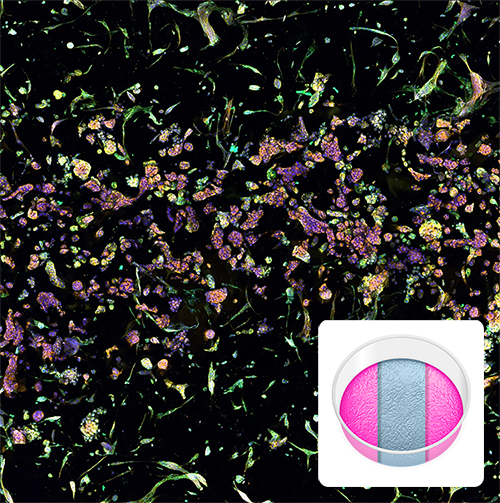
Triple Matrix Model
A triple-matrix design for recapitulating tissue architecture and studying complex cell interactions. Compatible with 96-well plates on RASTRUM and RASTRUM Allegro.
Applications
- Migration and invasion
- Tissue architecture recapitulation
- Cell signaling and interaction
- Wound healing and gap closure assays
Features
- Co-culture three distinct cell types in side-by-side compartments
- Independently define matrix stiffness, composition, and cellular components
- Visualize cell movement and interaction between compartments
- Simulate gap closure for wound healing and migration studies
Image illustrates tumor-fibroblast cell interactions within RASTRUM’s Triple Matrix Model architecture. Normal human lung fibroblasts (NHLFs) printed in the top and bottom matrix compartments of the model architecture interact with A549 lung adenocarcinoma cells growing within the middle compartment. Cells were labelled using the PhenoVue Cell Painting Kit: nuclei (blue, Hoechst), endoplasmic reticulum (green, concanavalin A), and actin (yellow , phalloidin), at day 7 post-printing. Inset: Triple Matrix Model architecture illustration.
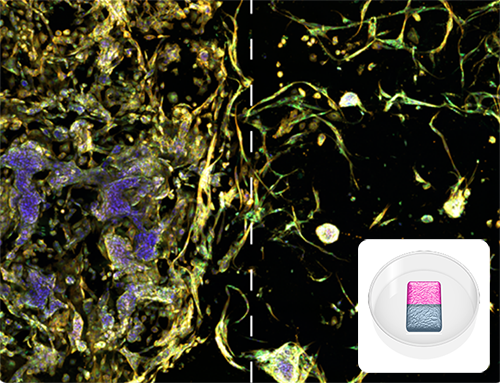
Dual Matrix Model
A dual-matrix architecture optimized for recapitulating tissue architecture and studying cellular interactions within a 3D environment. Compatible with 96-well plates on RASTRUM and RASTRUM Allegro.
Applications
- Migration and invasion studies
- Tissue architecture recapitulation
- Cell signaling and interaction
Features
- Enables imaging of cell signaling and interaction with clarity
- Provides 3D models with defined and reproducible tissue architecture
- Facilitates advanced studies on cellular behavior in complex environments
Image illustrates tumor microenvironment with spatial separation in RASTRUM’s Dual Matrix Model architecture. Lung cancer (A549) cells (left) and normal human lung fibroblast (right) were printed in a Dual Matrix Model architecture on RASTRUM and imaged after seven days in vitro. Dotted lines indicate the boundaries between the matrices. Cells were labelled using the PhenoVue Cell Painting Kit: nuclei (blue, Hoechst), endoplasmic reticulum (green, concanavalin A), and actin (yellow , phalloidin). Inset: Dual Matrix Model architecture illustration.
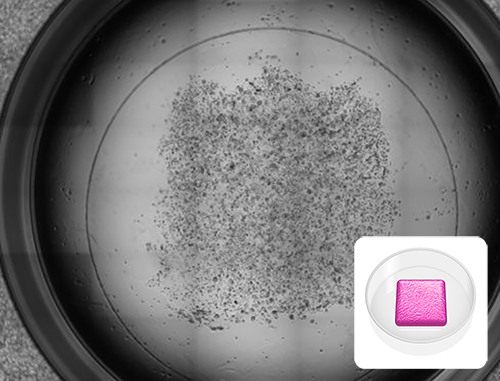
Removable Model
A large removable architecture designed for 24-well plates and printed on coverslips, offering exceptional versatility for implantation and downstream analyses. Compatible with 24-well plates on RASTRUM and RASTRUM Allegro.
Applications
- Implantation studies
- Immunohistochemistry
- Spatial biology
Features
- Easy removal for microscopic imaging
- Simplified handling for embedding and sectioning
- Ideal for workflows requiring intact tissue preparation
Image shows cancer cell culture in a 24-well plate format in preparation for immunohistochemistry. MCF-7 cells were printed in the RASTRUM Removable Model architecture and imaged after 3 days. The coverslip facilitates easy removal of the intact matrix for downstream applications. Inset: Removal Model architecture illustration.
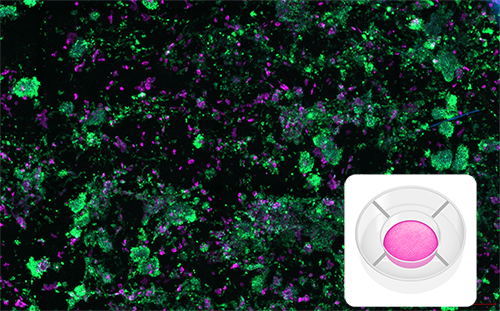
Mematix Model
A single-matrix architecture for transwell insert membranes, enabling advanced migration and signaling studies. Compatible with 96-well plates on RASTRUM and RASTRUM Allegro.
Applications
- Evaluation of cell movement and migration
- Paracrine signaling studies
Features
- Supports two separate media chambers for controlled chemokine or chemoattractant gradients
- Allows migration of cells through the membrane for advanced migration studies
Immuno-oncology model generated with RASTRUM, showcasing interactions between PBMCs and A549 cells in a Mematix Model architecture, imaged after 6 days. CD45+ immune cells (green) are observed interacting with Pan-CK+ cancer cells (magenta). Inset: Mematix Model architecture illustration.
How it works
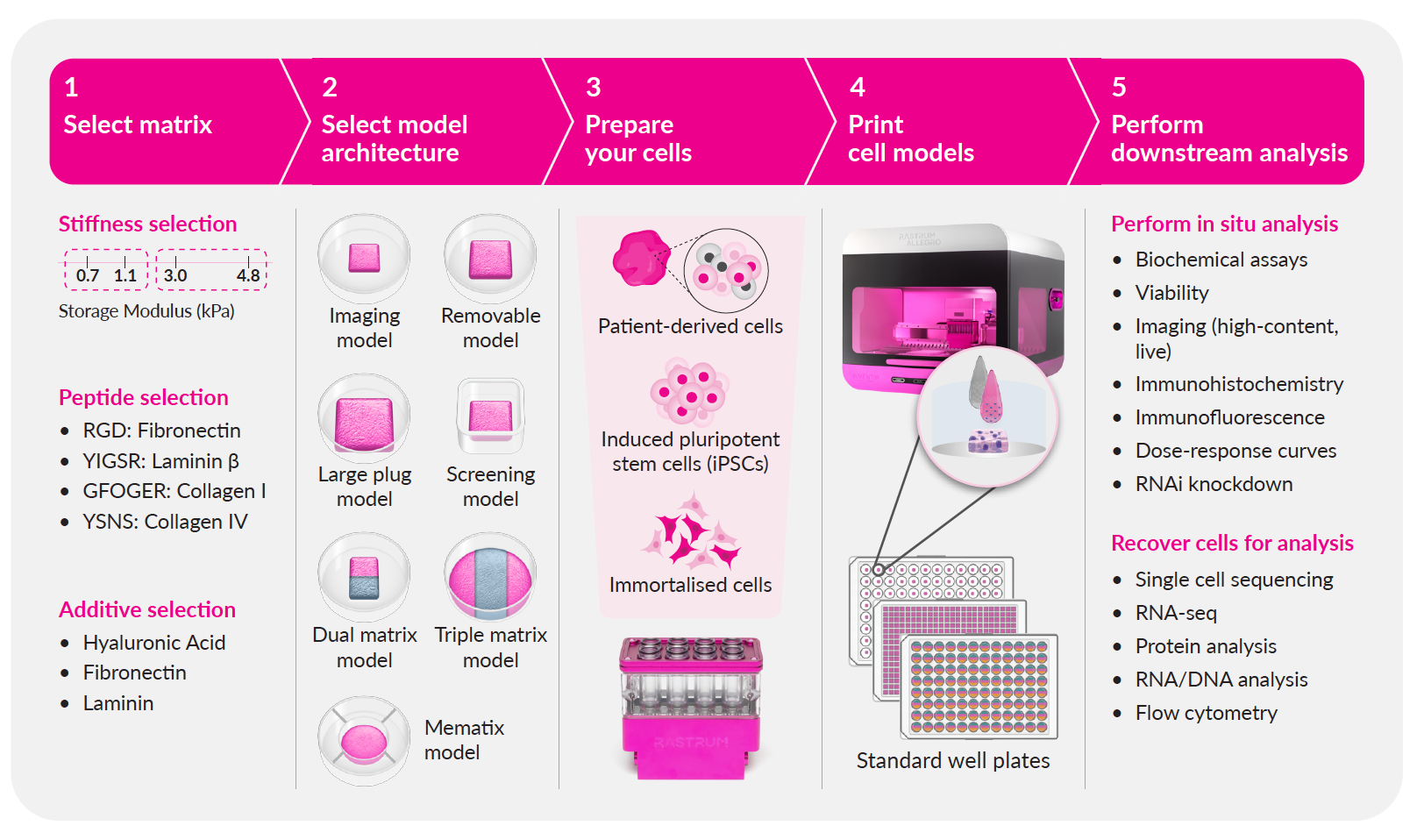
RASTRUM technology is designed to streamline your 3D cell model development from start to finish, delivering biologically relevant results with efficiency and precision. Simply point, click, and experiment with our cloud-based guided experimental design, ready-to-use protocols, and expert assistance.
- Select your matrix: Choose from our large library of flexible, tunable, xeno-free matrices to mimic the tissue microenvironment to suit your research.
- Select your cell model architecture: Choose the structure and configuration of your cell model from our options to suit your experimental needs.
- Prepare your cells: Follow a simple protocol to prepare your patient-derived, iPSC or immortalized cells
- Print your cell models: Utilize RASTRUM's drop-on-demand bioprinting technology to rapidly and reproducibly generate your 3D cell models.
- Perform your downstream analysis: Grow your cells and easily integrate cell model analysis with existing downstream workflows and readouts.


