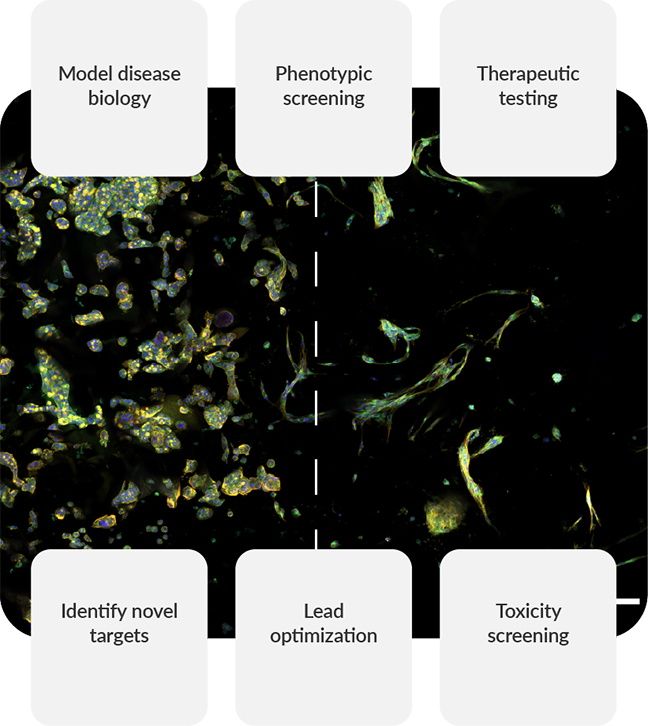
Accelerating drug discovery with 3D cell models
Breakthrough therapies start with the right model. The RASTRUM™ 3D cell culture platform empowers researchers to build high-throughput, reproducible, and biologically relevant cell models that accelerate drug discovery. By seamlessly integrating into screening workflows, RASTRUM Allegro enhances hit identification, optimizes lead selection, and improves translatability—bringing the next generation of medicines closer to reality.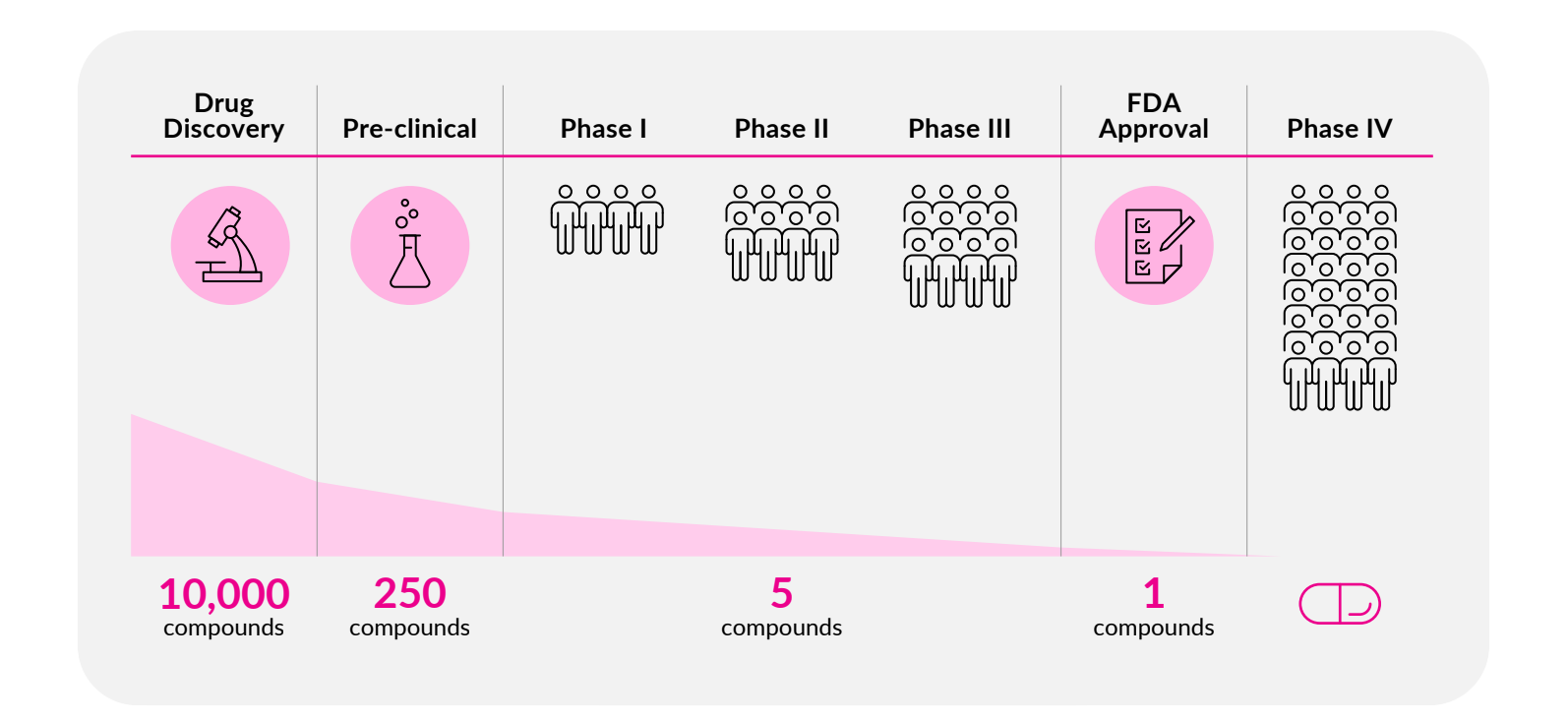
The challenge: Overcoming drug discovery bottlenecks
More than 90% of drug candidates fail during clinical development, often due to inadequate preclinical models that do not accurately predict human responses.
- 2D cultures lack the complex cell-cell and cell-matrix interactions necessary for capturing disease biology.
- Animal models are costly, time-consuming, and frequently fail to translate to human outcomes.
- Traditional approaches introduce variability, leading to inconsistent and unreliable screening results.
To bridge the gap, researchers need a phenotypic cell model approach—one that exhibits a clinically relevant phenotype, expresses disease-relevant molecular signatures, and is standardized, reproducible, and scalable for modern drug discovery.
Infographic highlighting the difficulty of getting compounds successfully through clinical trials to FDA approval.
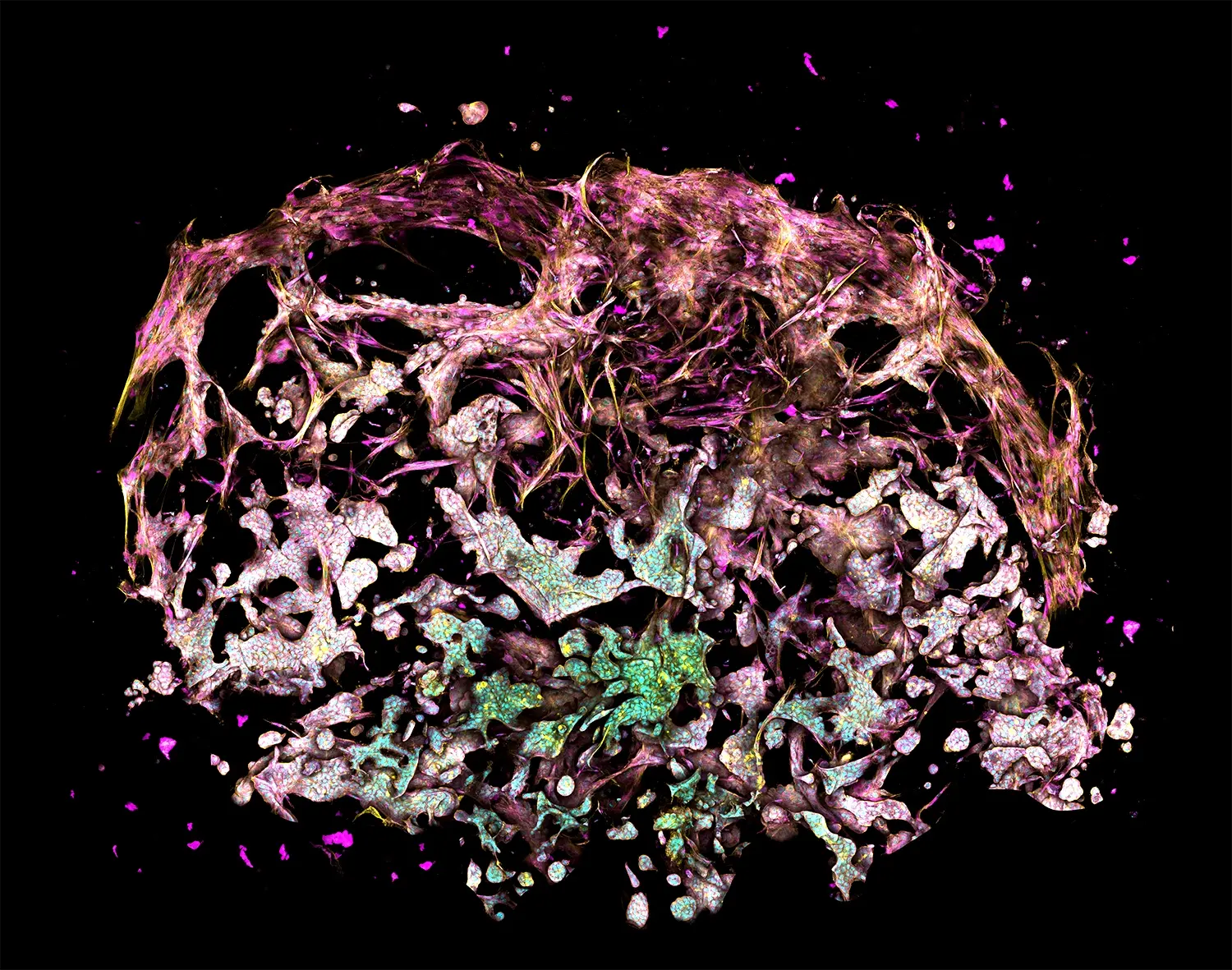
RASTRUM: Transforming drug discovery with advanced 3D cell models
RASTRUM Allegro enables the rapid creation of scalable, reproducible 3D cell models that better replicate human biology—providing researchers with a more predictive system for screening drug candidates, optimizing lead compounds, and improving clinical translation.
Why RASTRUM for drug discovery?
- Clinically relevant phenotypic models: Capture disease-specific molecular signatures and drug response behaviors.
- Advanced co-cultures: Build complex multi-cellular environments to study interactions between diseased and healthy tissues.
- Scalable and reproducible: High-throughput, standardized 3D models for consistent screening and validation.
- Seamless integration with drug screening workflows: Optimized for high-content imaging, automation, and screening pipelines.
RASTRUM Allegro empowers drug discoverers to advance phenotypic screening, lead optimization, and preclinical drug development—bridging the gap between early-stage research and clinical success.
The figure depicts a lung cancer tri-culture model made with RASTRUM Allegro. Primary normal human lung fibroblasts (NHLFs) were printed with primary Human Umbilical Vein Endothelial Cells (HUVECs) and A549 lung adenocarcinoma cells using RASTRUM’s Imaging Model architecture. Cells were cultured for 7 days and stained using a PhenoVue Cell Painting Kit (Revvity). Cell nuclei (cyan) were labeled with Hoescht, filamentous actin (yellow) with phalloidin, and membrane glycoproteins (magenta) with concanavalin A.
3D cell model applications in drug discovery
Disease modeling for target identification
Understanding disease mechanisms at a cellular level is essential for discovering new therapeutic targets and developing effective treatments. RASTRUM Allegro enables researchers to engineer biologically relevant 3D models for deeper insights into disease pathology.
- Recreate disease-specific microenvironments: Model patient-relevant tissue structures to uncover novel drug targets.
- Validate mechanisms of action: Study how drug candidates interact with native-like 3D cellular environments.
- Improve model consistency: Reduce experimental variability with automated, reproducible 3D cell culture.
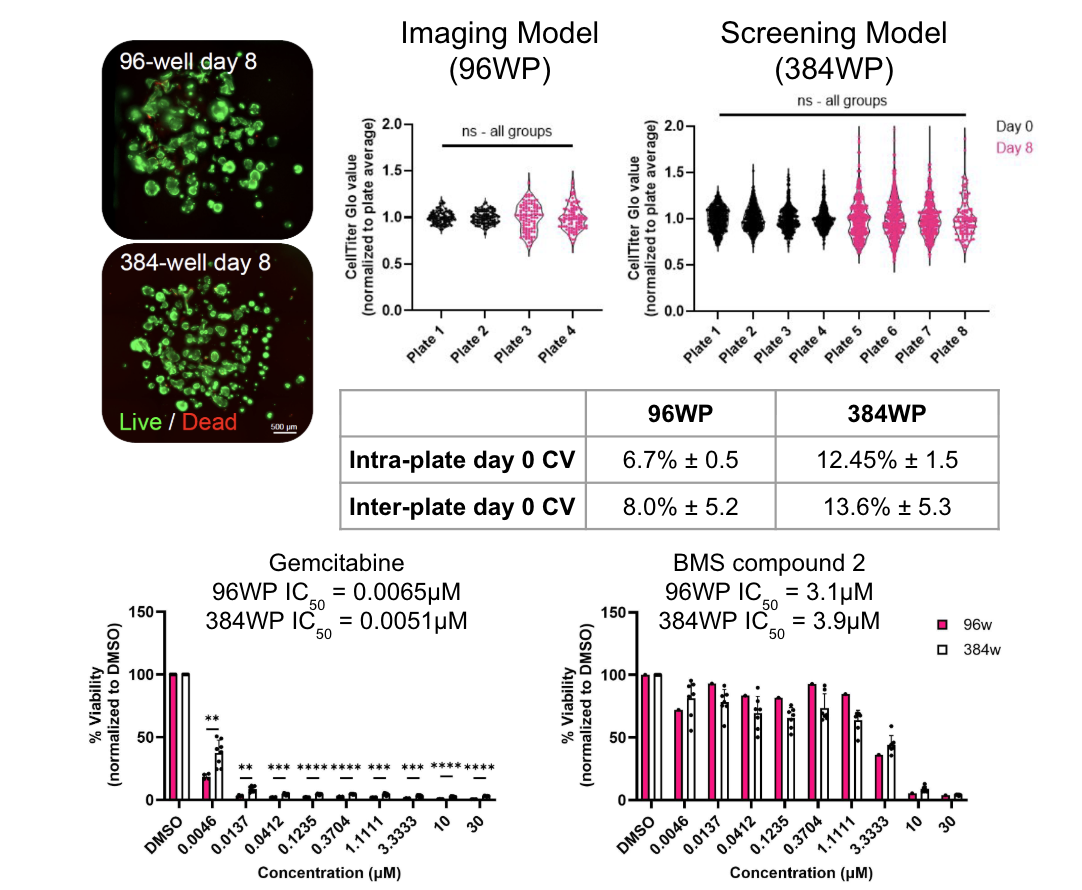
The figure depicts the development of a reproducible, high-throughput, and screenable 3D PDAC model using the RASTRUM Allegro platform. Viability of PDAC monocultures post-printing and at endpoint. Green = calcein-AM (live cells), red = EthD-III (dead cells). Scale bar = 500 μm. Reproducibility of PDAC culture viability across multiple plates and formats. Intra- and inter-plate coefficient of variation (CV) was calculated using CellTiter-Glo measurements (n=96–384 wells/plate; N=2–4 plates/format). Error = standard deviation (SD). Drug responses in PDAC monocultures printed into different plate formats. Viability was normalized to DMSO controls. n=6 wells per treatment condition; N=8 plates per compound (384w), or 1–4 plates/compound (96w). Each data point ( ) represents the average of replicate wells from a separate plate. Error bars = SD.
Phenotypic screening & lead optimization with 3D cell models
Capturing disease-relevant phenotypes is critical for identifying high-value drug candidates and optimizing lead compounds. RASTRUM Allegro enables the creation of high-throughput, reproducible 3D cell models that enhance the predictive power of phenotypic drug screening.
- Improve translatability in drug discovery: Develop 3D cell models that express clinically relevant molecular signatures, improving the accuracy of lead selection.
- Model complex disease phenotypes: Incorporate key cell-cell and cell-matrix interactions to study drug effects in a biologically meaningful context.
- Standardized, scalable drug screening: Generate consistent, assay-ready 3D cultures optimized for high-content imaging and automated workflows.
The figure depicts a model of the tumor-stroma interface in a Triple Matrix Model made with RASTRUM. Fibroblasts (primary human lung fibroblasts) and endothelial cells (primary HUVECs) were grown next to lung adenocarcinoma cancer cells (A549) and fibroblasts (HUVECs) were cultured for 10 days. Endothelial cells were stained with CD31 (yellow) and fibroblasts and endothelial cells were stained with Phalloidin (blue).
Oncology drug development & tumor microenvironment studies
Cancer drug discovery requires models that accurately reflect tumor heterogeneity and microenvironmental factors that drive treatment response and resistance. RASTRUM Allegro enables the development of advanced 3D tumor models that improve the clinical relevance of oncology drug testing.
- Engineer complex tumor microenvironments: Incorporate immune cells, fibroblasts, and extracellular matrix components for realistic tumor modeling.
- Study drug resistance and treatment response: Analyze how tumor-stroma interactions impact drug efficacy.
- Optimize targeted therapies: Test small molecules, biologics, and immunotherapies in standardized 3D cancer models.
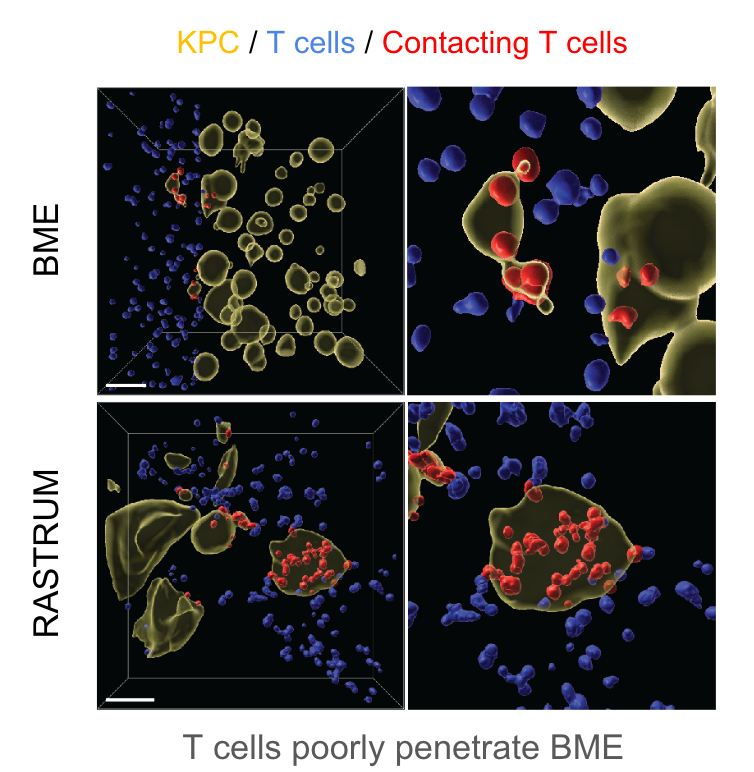
The figure depicts pancreatic cancer cells cultured in 4.8kPa matrices for 3 days before 12,000 activated cytotoxic T lymphocytes (CTLs) were added to the media in each well. Imaging and Imaris analysis shows CTL infiltration 2 days post-addition and allows quantification of interactions (red) between cancer cells (yellow) and T cells (blue) in RASTRUM matrices and basement membrane extract (BME).
Toxicology & drug safety testing with predictive 3D cell models
Traditional toxicity testing often fails to predict human responses, leading to late-stage failures in drug development. RASTRUM Allegro enables scalable, biologically relevant 3D models that improve drug safety assessments.
- Assess drug toxicity in a human-relevant system: Evaluate off-target effects, metabolism, and dose responses in reproducible 3D models.
- Reduce reliance on animal testing: Use advanced cell-based assays to generate more predictive toxicity data.
- Enhance preclinical screening efficiency: Standardized 3D models ensure reliable, high-throughput safety testing.
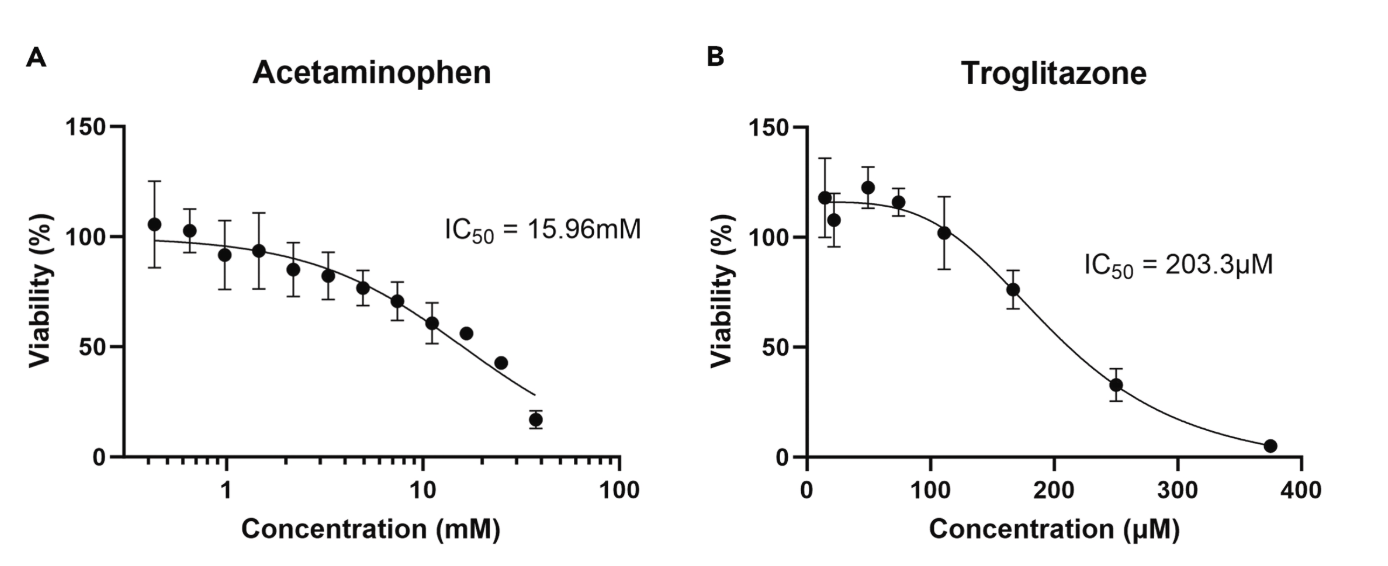
Image illustrates drug responsiveness testing in RASTRUM primary human hepatocyte 3D cell models. (A) Acute drug exposure to a widely used pain reliever/analgesic in RASTRUM-printed PHH achieved an IC50 comparable to other 3D PHH models. Measured by CellTiter-Glo 3D at day 5 post-printing, after 2 days (48 h) of exposure to the analgesic. Data points are means ± SDs from n = 3 technical replicates per concentration. (B) Long-term viability of PHH in RASTRUM Matrix enabled chronic drug toxicity studies with a hepatotoxic reference compound. Measured by CellTiter-Glo 3D at day 10 post-printing and after 7 days of exposure to the hepatotoxic reference compound. Data points are means ± SDs from n = 3 technical replicates per concentration. Figure 4 in the STAR Protocol: Protocol to create phenotypic primary human hepatocyte cultures using the RASTRUM 3D cell model platform
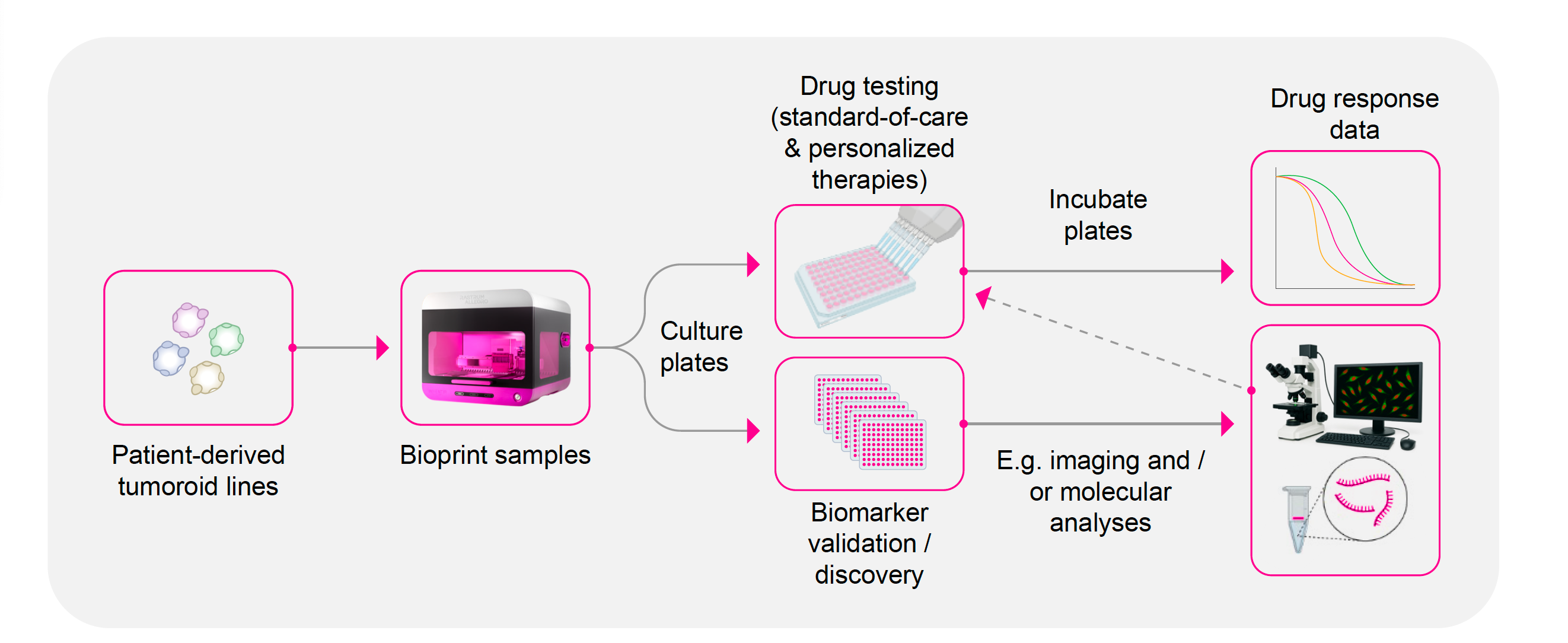
Combination therapy & personalized medicine approaches
Testing drug combinations using patient-derived models is critical for advancing precision medicine and personalized treatment strategies. RASTRUM Allegro allows researchers to evaluate drug synergies and patient-specific responses in highly controlled 3D cultures.
- Develop patient-derived models for personalized treatment: Build models using patient-specific cells to guide decisions.
- Support precision medicine initiatives: Generate clinically relevant, reproducible models that align with personalized drug screening strategies.
- Test combination therapies with real-world complexity: Study how multiple drugs interact in dynamic 3D cell models.
The infographic illustrates an example of how RASTRUM Allegro seamlessly integrates into translational pipelines in the precision medicine space for drug testing of standard of care and personalized therapies and biomarker validation and discovery work.
FAQs
RASTRUM Allegro is designed to seamlessly integrate with high-content imaging, automated liquid handling, and multi-well plate screening workflows. Its standardized 3D cultures are compatible with fluorescence, luminescence, and viability assays, ensuring effortless adoption into existing drug screening and preclinical research pipelines.
RASTRUM Allegro is for research use only; however, researchers are actively using it in preclinical studies to develop patient-derived models, evaluate drug response variability, and explore precision medicine approaches. By enabling highly controlled, reproducible 3D cultures, it supports the development of more targeted and effective therapies.
2D cultures lack key cell-cell and cell-matrix interactions, failing to replicate native tissue structure or accurately predict in vivo drug responses. RASTRUM Allegro-generated 3D models provide greater biological relevance, capturing disease-specific phenotypes, signaling pathways, and drug resistance mechanisms, leading to more reliable and clinically translatable results.
RASTRUM Allegro enables fully defined, animal-free 3D cell culture workflows, eliminating the need for animal-derived matrices like Matrigel. By providing highly controlled, reproducible 3D models, RASTRUM helps researchers reduce reliance on animal models while improving the accuracy of preclinical drug testing.
Yes, RASTRUM Allegro was designed with automation in mind. It seamlessly integrates with liquid handling systems, robotic workstations, and high-throughput screening platforms. For researchers with specific automation needs, we work closely with organizations to ensure RASTRUM Allegro fits into their existing workflows, enabling efficient, scalable 3D cell culture integration into drug discovery pipelines.