
Biologically matched matrices for 3D cell models
RASTRUM™ Matrices are designed to provide a biologically relevant extracellular matrix (ECM)-like environment for building reliable and reproducible 3D cell models. With tunable properties, xeno-free composition, and support for a wide range of cell types, these matrices enable researchers to advance drug discovery and disease modeling with confidence.


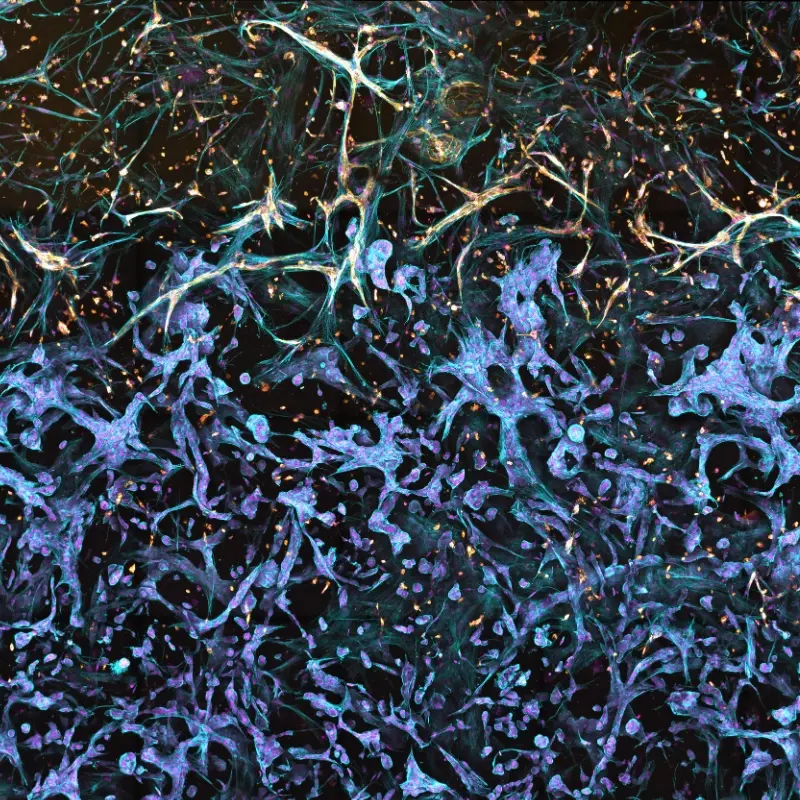
Build a biologically relevant tissue microenvironment
Cells don’t exist in isolation—they interact with their extracellular matrix and surrounding cells in complex ways. RASTRUM Matrices enable the production of spatially defined 3D tissue architectures that replicate native microenvironments, from tumor-stroma interactions to immune cell infiltration. By controlling both matrix composition and spatial organization, researchers can study disease mechanisms and therapeutic responses with unprecedented reproducibility.
The image depicts an in vitro model of the tumor-stromal interface made with RASTRUM. A co-culture of primary normal human lung fibroblasts (NHLF cells) and primary human vascular endothelial cells (HUVECs) were printed to interface with A549 lung adenocarcinoma cells, using a triple matrix model architecture. Cell nuclei (purple) were labeled with DAPI, filamentous actin (blue) with phalloidin, and the endothelial marker CD31 (gold) with a protein-specific antibody, at day 9 post-printing.
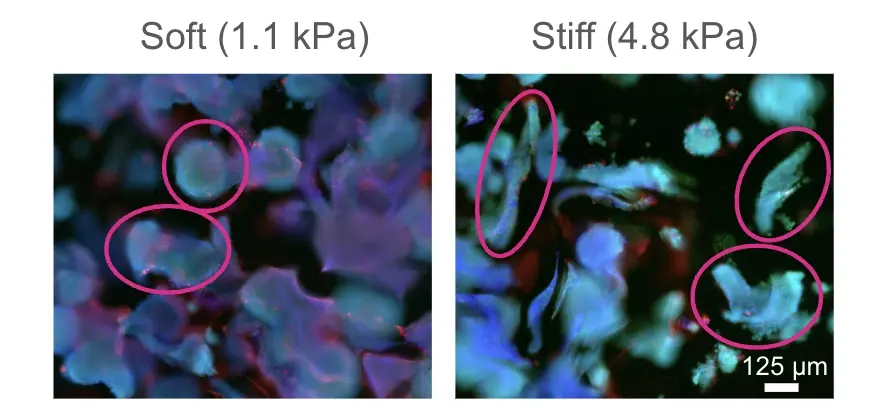
Fine-tune your model: Modulate matrix stiffness
The mechanical properties of the ECM directly influence cell behavior, affecting processes like differentiation, migration, and tissue development. With RASTRUM Matrices, stiffness can be precisely adjusted, allowing researchers to mimic the biomechanical properties of native tissues—from the softness of brain tissue to the rigidity of fibrotic tumors. This tunability enables more physiologically relevant disease modeling and a deeper understanding of mechanotransduction in cell biology, and expands cell compatibility by accommodating cell types that require specific microenvironments, such as pancreatic cells needing higher stiffness.
The image depicts the modulation of the tumor microenvironment stiffness in an ovarian cancer model made with RASTRUM. A culture of A2780 ovarian cancer immortalized cells were grown in 1.1 or 4.8 kPA matrices. Cell nuclei (blue) were labeled with DAPI, live cells (green) were labeled with calcein, and dead cells (red) labeled with ethidium homodimer-3, at day 7 post-printing. A change in shape of the spheroids from round to elongated was observed in the stiffer matrix.
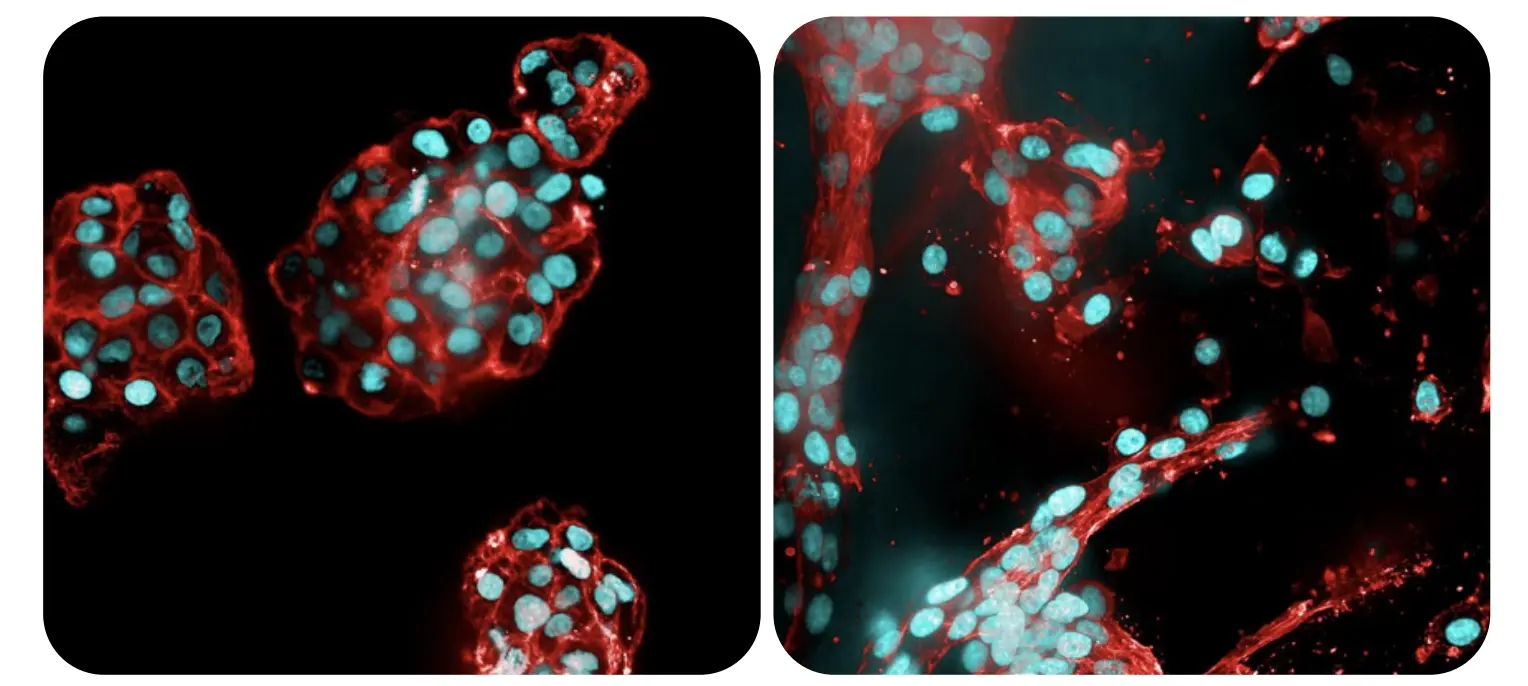
Customizable microenvironments: Tailored adhesion peptides and additives
Beyond mechanical properties, the biochemical composition of the ECM plays a crucial role in supporting cellular function. RASTRUM Matrices support the incorporation of key ECM proteins and glycosaminoglycans to fine-tune cell attachment, survival, and specialized functions. Researchers can customize their matrices with adhesion peptides (Fibronectin, Laminin β, Collagen I, Collagen IV) and full-length ECM components (Hyaluronic acid, Laminin-521, Laminin-211, Fibronectin) to enhance tissue-specific behaviors and cell-matrix interactions.
The image depicts glioblastoma cells grown in a RASTRUM model. U-87 glioblastoma cells were grown in RASTRUM Matrix in the absence (left) or presence (right) of Fibronectin adhesion peptide (RGD). Cell nuclei (blue) were labeled with DAPI and filamentous actin (red) were labeled with phalloidin (green). The presence of RGD in the matrix promotes a branched and spindle-like morphology, which represents the clinically relevant invasive phenotype for glioblastoma.
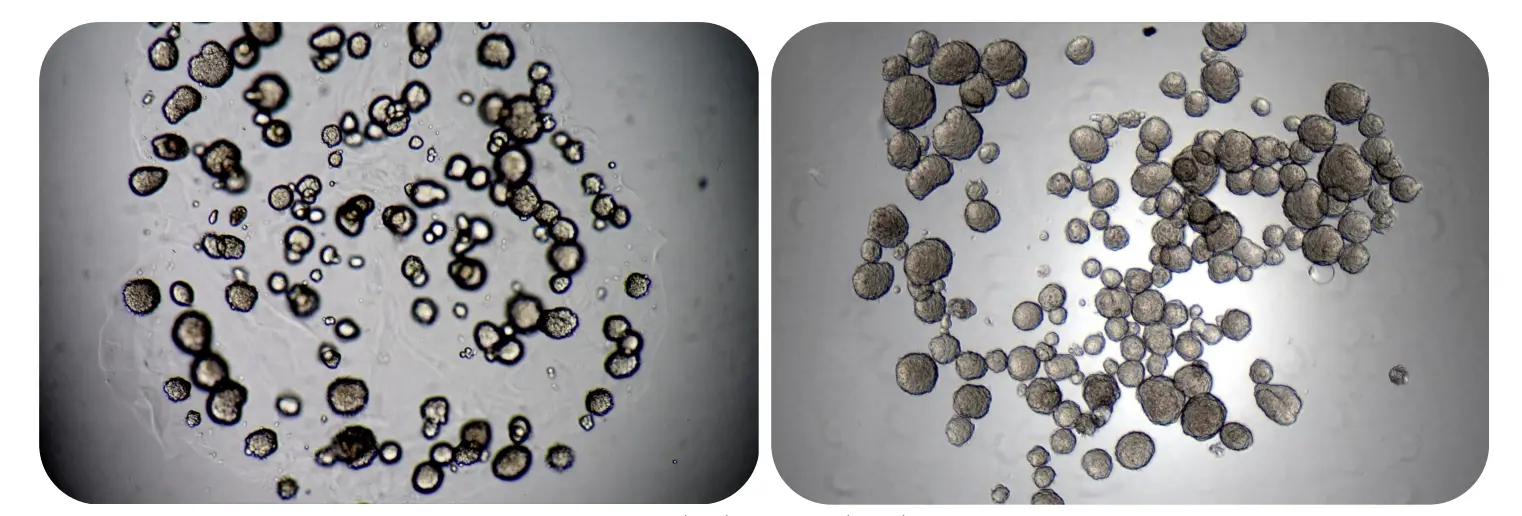
Dynamic and accessible matrices: Supporting cell behavior and cell recovery
RASTRUM Matrices are designed to evolve with cells, featuring MMP-sensitive sites that enable cell-secreted proteases to remodel the matrix—a key characteristic of native ECM dynamics. This allows cells to migrate, interact with neighboring cells, and modify their microenvironment, better simulating in vivo behavior. The porous structure facilitates immune cell infiltration, cell-cell communication, and the diffusion of bioactive molecules, ensuring a physiologically relevant research model. When analysis is needed, the matrix is designed for seamless cell recovery, with RASTRUM’s Cell Recovery Solution enabling downstream applications such as single-cell sequencing, flow cytometry, and more.
The image depicts tumor spheroids in RASTRUM Matrix before (left) and after (right) adding RASTRUM Cell Recovery Solution. Once the RASTRUM Matrix is degraded, the released spheroids are visible in a single focal plane at the bottom of the well.
Xeno-free matrices for reproducibility and consistency
Traditional ECM-based hydrogels often contain animal-derived components, leading to batch variability and unpredictable experimental outcomes. RASTRUM Matrices provide a fully synthetic, PEG-based alternative, ensuring batch-to-batch consistency and enhanced reproducibility. By minimizing reliance on animal-derived ECM components, researchers can achieve greater control over their 3D cell culture conditions, leading to more reliable and reproducible results.
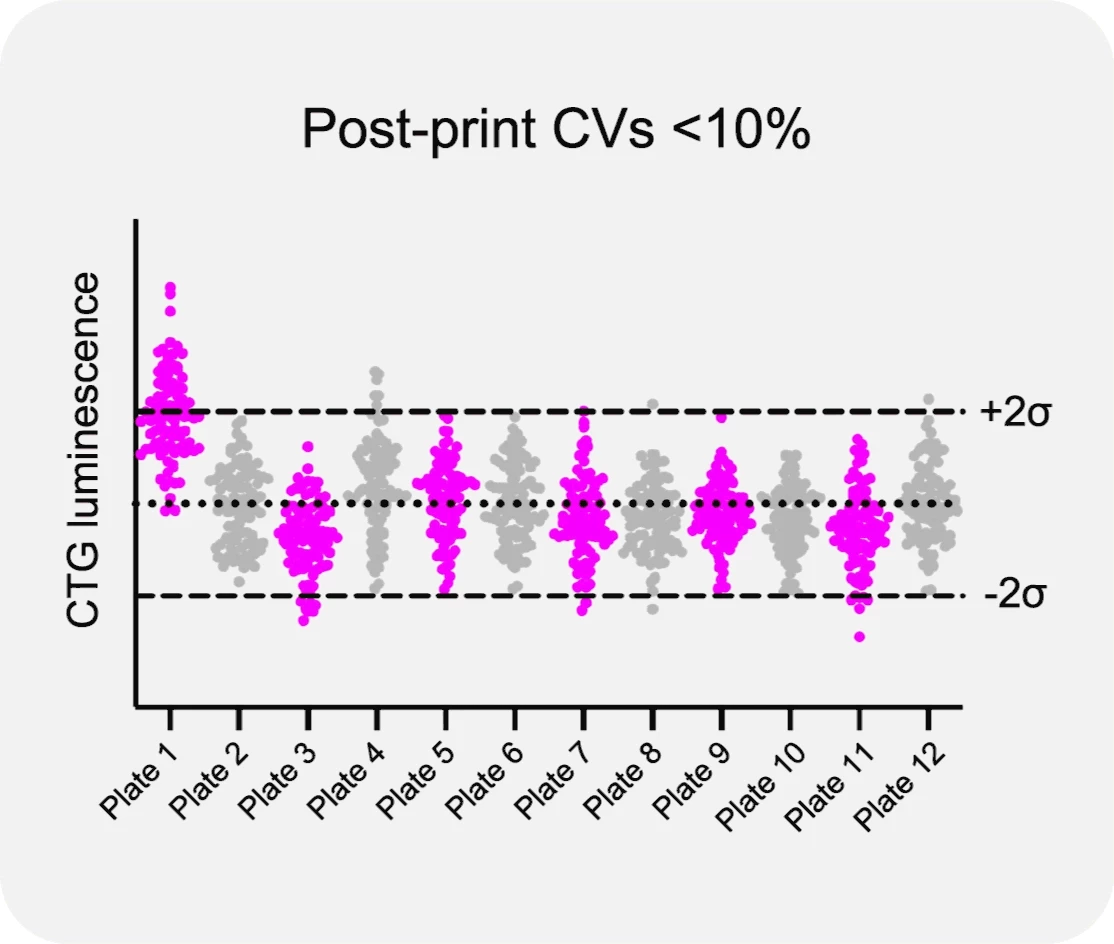
The consistency of RASTRUM Allegro in high-throughput screening applications is demonstrated in the figure, showing MCF-7 cells and liver cancer (HepG2) cells printed with Imaging Models across twelve 96-well plates. Post-print analysis demonstrates intra-plate and inter-plate CV values <10%. MCF-7 cells printed as Screening Models in twelve 384-well plates with treatment with paclitaxel chemotherapy drug. Analysis post-treatment demonstrates intra-plate and inter-plate CV values <19%.
True 3D culture: Preventing 2D monolayer formation
Cells in 3D culture often migrate toward plastic surfaces, where they attach, spread, and lose their 3D characteristics—leading to unwanted activation and skewed experimental results. RASTRUM overcomes this challenge with an inert base layer that coats the bottom of the well, preventing direct contact with the plastic and ensuring cells remain suspended within the matrix. This innovation maintains the integrity of the 3D microenvironment, preventing the formation of 2D monolayers and enabling more physiologically relevant experiments.
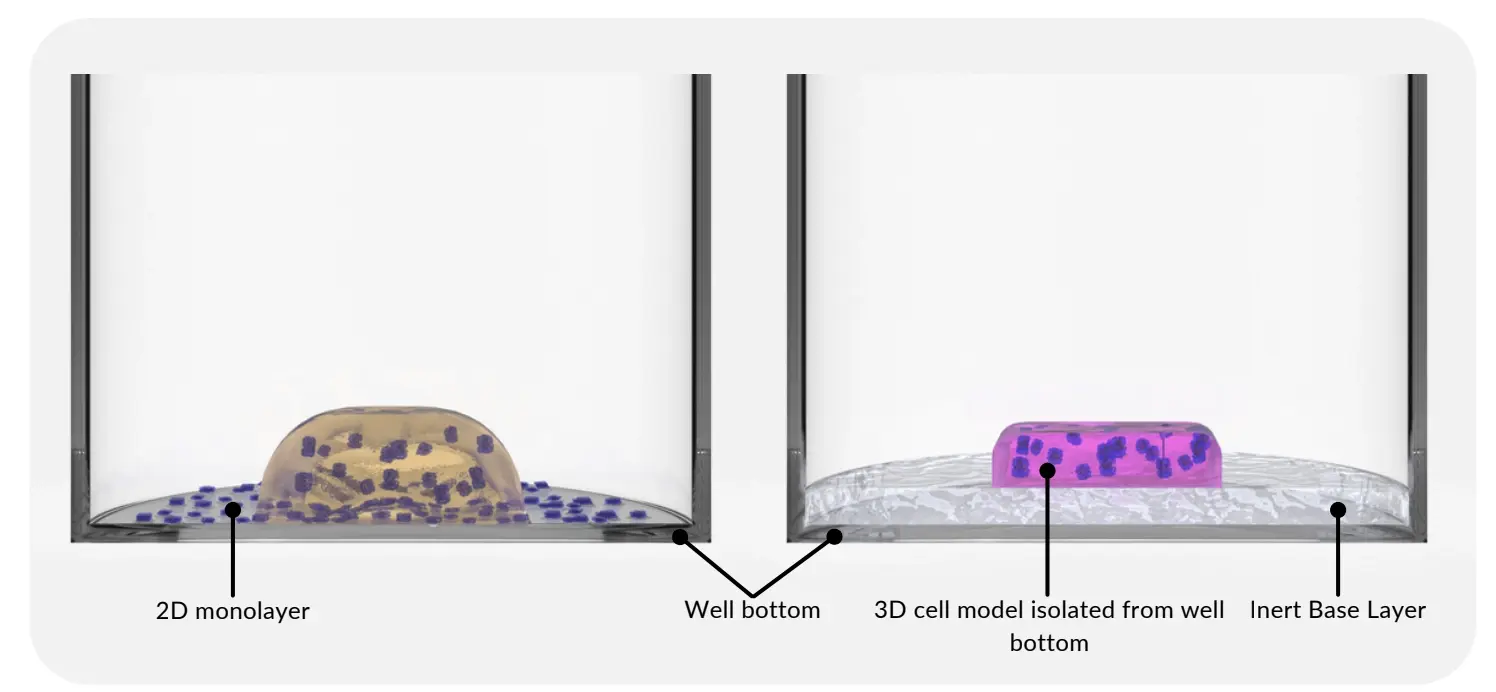
The image depicts 2D monolayer formation in wells containing 3D cell models made using basement membrane extract (left). RASTRUM 3D cell models have an Inert Base Layer that prevents 2D monolayer formation (right).
How RASTRUM works
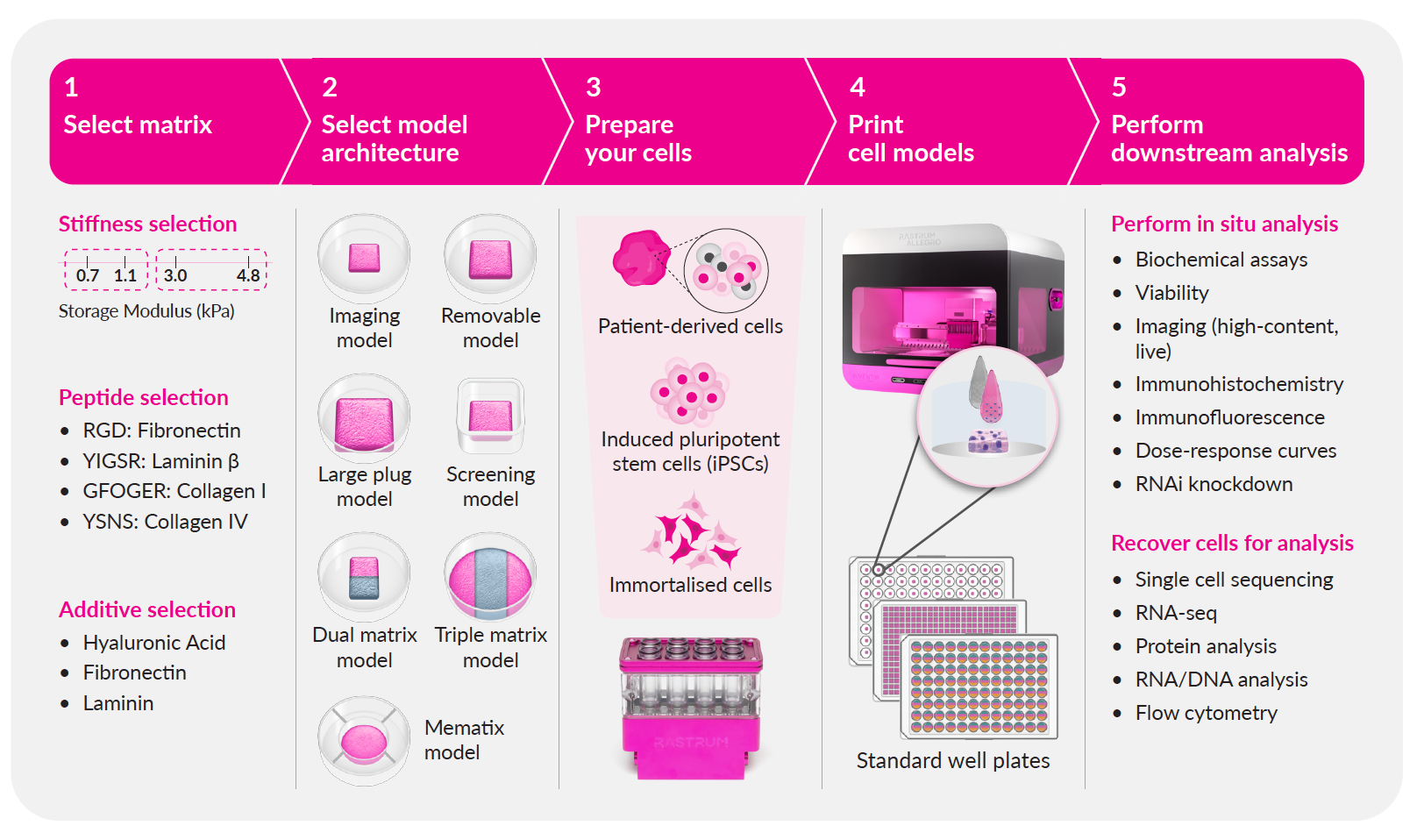
RASTRUM technology is designed to streamline your 3D cell model development from start to finish, delivering biologically relevant results with efficiency and precision. Simply point, click, and experiment with our cloud-based guided experimental design, ready-to-use protocols, and expert assistance.
- Select your matrix: Choose from our large library of flexible, tunable, xeno-free matrices to mimic the tissue microenvironment to suit your research.
- Select your cell model architecture: Choose the structure and configuration of your cell model from our options to suit your experimental needs.
- Prepare your cells: Follow a simple protocol to prepare your patient-derived, iPSC or immortalized cells
- Print your cell models: Utilize RASTRUM's drop-on-demand bioprinting technology to rapidly and reproducibly generate your 3D cell models.
- Perform your downstream analysis: Grow your cells and easily integrate cell model analysis with existing downstream workflows and readouts.
Resources to empower your research
 Beyond simple co-cultures: Development of complex tissue architectures in 3D
This technical note will help you explore the tissue microenvironment with 3D culture.
Beyond simple co-cultures: Development of complex tissue architectures in 3D
This technical note will help you explore the tissue microenvironment with 3D culture.
 Printable and tunable 3D cell culture environments using RASTRUM™ Matrices
Read this application note to explore how you can tailor 3D cell culture environments with tunable matrices
Printable and tunable 3D cell culture environments using RASTRUM™ Matrices
Read this application note to explore how you can tailor 3D cell culture environments with tunable matrices
 The tumour microenvironment in phenotypic drug discovery
Explore key considerations for using 3D cell models in phenotypic drug discovery
The tumour microenvironment in phenotypic drug discovery
Explore key considerations for using 3D cell models in phenotypic drug discovery


