Advancing personalized medicine with patient-derived 3D cell models
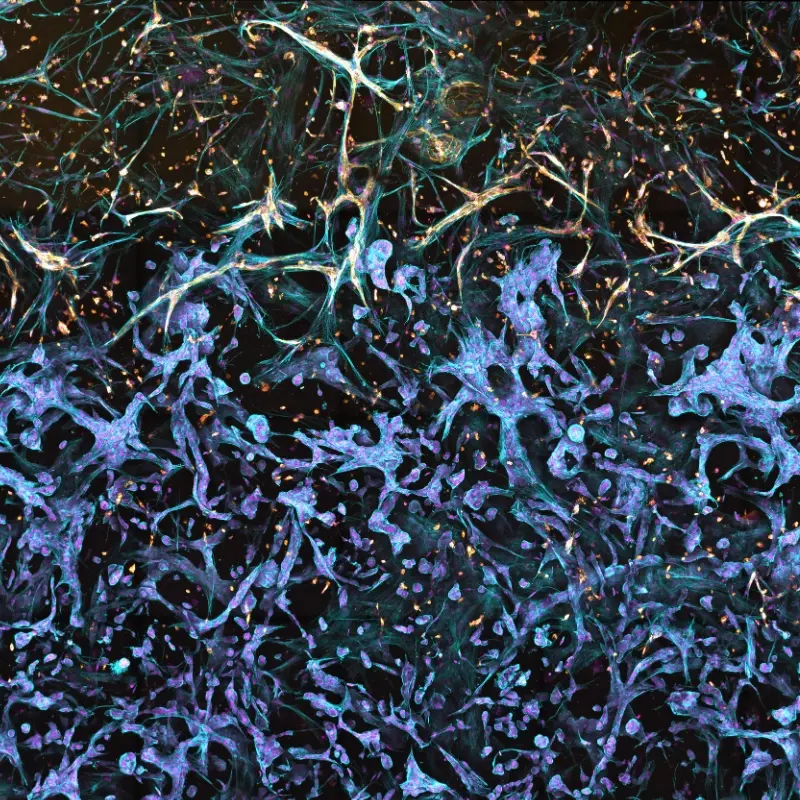
Scalable, reproducible, and biologically relevant. RASTRUM™ Allegro enables researchers to build 3D cell models from patient- and iPSC-derived sources, preserving phenotype and accelerating translational insights in oncology, neuroscience, and beyond.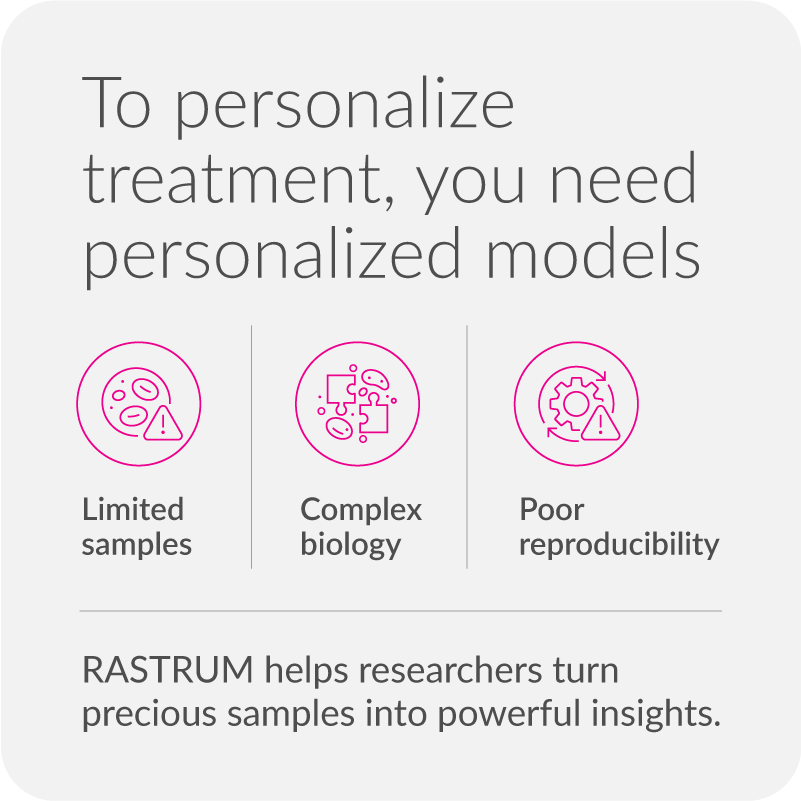
The challenge: Making personalized cell models meaningful and scalable
Advancing personalized and translational medicine requires more than patient-derived cells—it demands model systems that preserve biology, scale efficiently, and produce actionable insights. But many researchers face common barriers:
- Limited and precious samples: Low cell yields from iPSC or patient-derived sources make it difficult to run multiple conditions or repeat experiments.
- Capturing complex biology: Current models often fail to reflect patient heterogeneity or maintain biomarker expression—undermining disease modeling and therapeutic relevance.
- Poor reproducibility: Manual workflows introduce inconsistency, making it hard to generate reproducible results across plates, experiments, or sample cohorts.
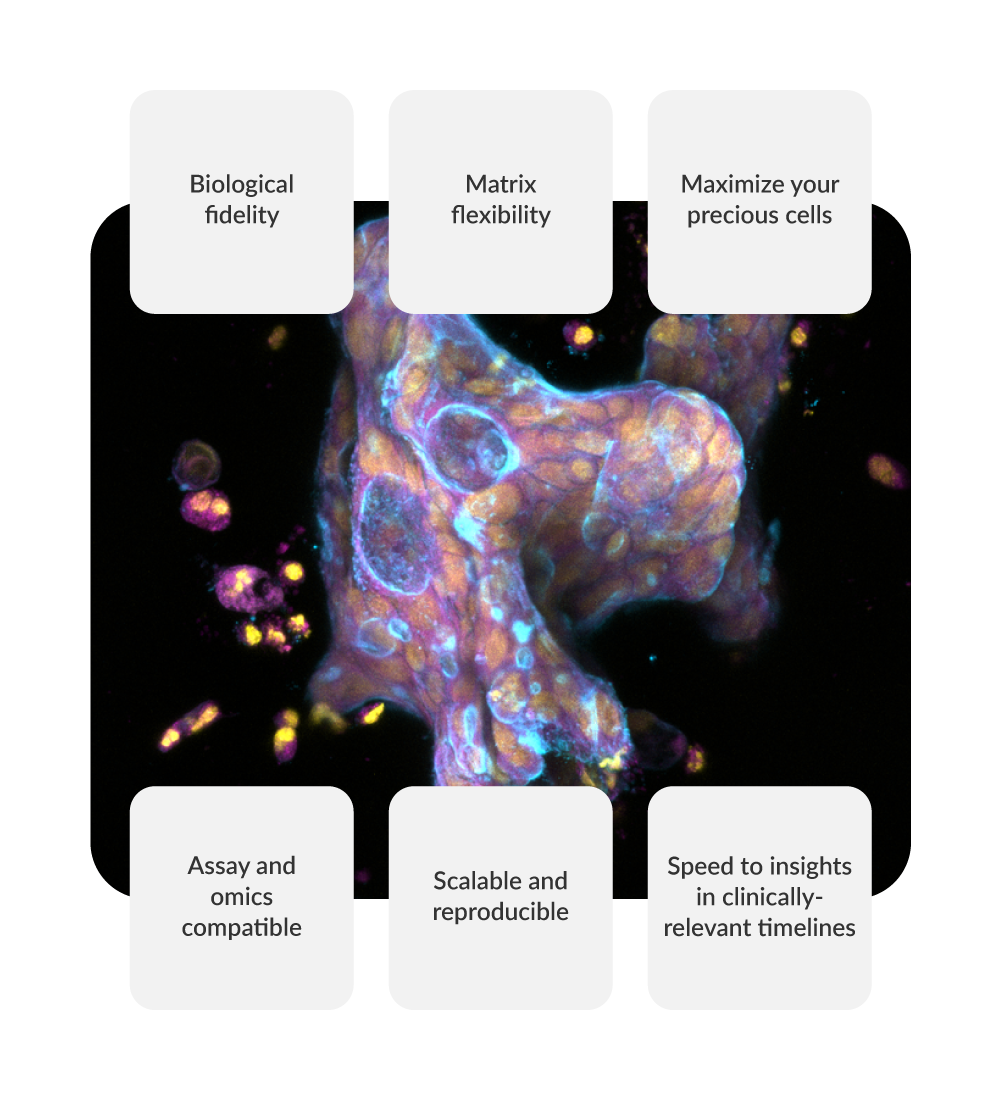
RASTRUM: Purpose-built for personalized and translational medicine
RASTRUM Allegro enables researchers to create scalable, reproducible 3D cell models from patient- and iPSC-derived cells—unlocking the full potential of personalized biology. Whether you're modeling disease heterogeneity or screening across patient subtypes, RASTRUM Allegro bridges the gap between complex biology and practical workflows.
Key advantages:
- Biological fidelity: Maintain disease-relevant gene expression and phenotypic markers throughout culture—enabling deeper, more reliable biological insights.
- Matrix flexibility: Tune biochemical and mechanical properties to mimic the microenvironmental signals that drive cell behavior—supporting disease-specific differentiation, function, and long-term viability.
- Maximize your precious cells: Conserve rare or low-yield samples while running multi-condition experiments in scalable 3D formats.
- Assay and omics compatible: Generate assay-ready 3D cultures optimized for imaging, transcriptomics, and functional profiling.
- Scalable and reproducible: Generate consistent, assay-ready 3D models across plates and conditions—built for high-throughput discovery.
The figure depicts a patient-derived colorectal cancer tumoroid. Primary colorectal cancer cells were printed using the RASTRUM Allegro Screening Model and cultured for 3 days. Tumoroids were then fixed and stained, with cell nuclei (yellow) labeled with Hoescht, filamentous actin (cyan) marked by phalloidin, and mitochondria (magenta) shown by PhenoVue Mitochondrial Stain.
3D Cell Model Applications in Drug Discovery
Modeling patient-specific disease biology
Capture heterogeneity. Preserve complexity.
RASTRUM Allegro enables researchers to build biologically relevant 3D models that reflect the molecular signature and tissue microenvironment of real patient tissues. From tumor-stroma dynamics to neuron–glia interactions, the platform supports co-culture models that recreate disease-relevant biology across a range of indications.
- Model patient-derived or iPSC-based systems with confidence
- Maintain biomarker expression and functional phenotype over time
- Customize matrix composition to reflect target tissue environments
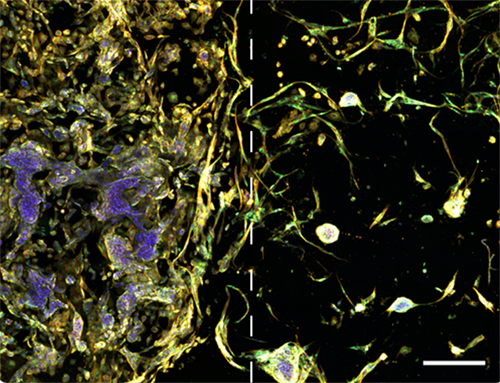
Image illustrates tumor microenvironment with spatial separation in RASTRUM’s Dual Matrix Model architecture. Lung cancer (A549) cells (left) and normal human lung fibroblast (right) were printed in a Dual Matrix Model architecture on RASTRUM and imaged after seven days in vitro. Dotted lines indicate the boundaries between the matrices. Cells were labelled using the PhenoVue Cell Painting Kit: nuclei (blue, Hoechst), endoplasmic reticulum (green, concanavalin A), and actin (yellow, phalloidin).
Scaling functional drug testing
Turn rare samples into reproducible data.
RASTRUM Allegro makes it possible to scale 3D models from limited patient material, enabling high-throughput, multi-condition screens across cell types and disease states. With this streamlined workflow, researchers can produce consistent, assay-compatible 3D models for functional drug testing—reflecting real-world biology at scale
- Use fewer cells to test more conditions
- Support multiplexed imaging, viability, and transcriptomics
- Compare therapeutic responses across diverse patient models
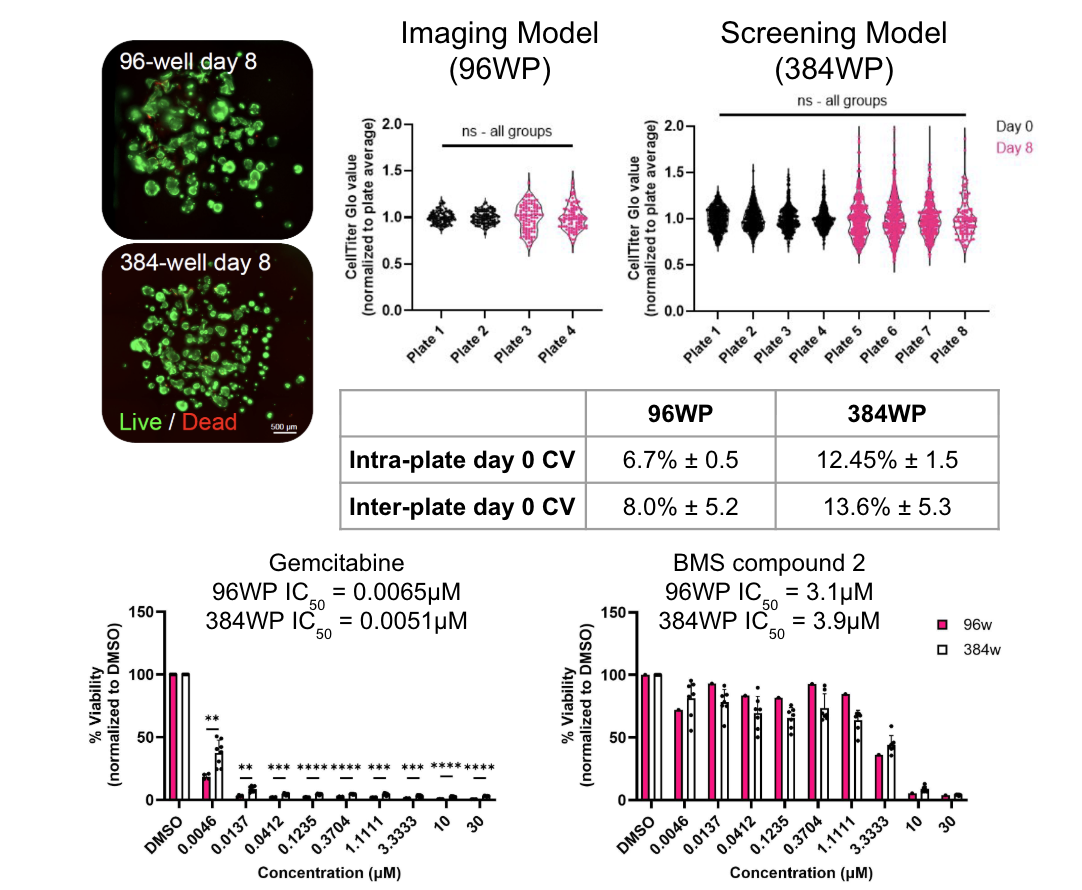
The figure depicts the development of a reproducible, high-throughput, and screenable 3D PDAC model using the RASTRUM Allegro platform. Viability of PDAC monocultures post-printing and at endpoint. Green = calcein-AM (live cells), red = EthD-III (dead cells). Scale bar = 500 μm. Reproducibility of PDAC culture viability across multiple plates and formats. Intra- and inter-plate coefficient of variation (CV) was calculated using CellTiter-Glo measurements (n=96–384 wells/plate; N=2–4 plates/format). Error = standard deviation (SD). Drug responses in PDAC monocultures printed into different plate formats. Viability was normalized to DMSO controls. n=6 wells per treatment condition; N=8 plates per compound (384w), or 1–4 plates/compound (96w). Each point represents the average of replicate wells from a separate plate. Error bars = SD.
Discovering personalized treatment responses
Reveal inter-patient variability. Unlock therapeutic insights.
RASTRUM Allegro supports systematic evaluation of how individual models respond to drugs, combinations, and targeted agents. Researchers can uncover subtype-specific vulnerabilities and identify biomarkers linked to response or resistance—laying the foundation for truly personalized therapeutic strategies.
- Interrogate treatment responses across patient-derived cohorts
- Profile drug response variability with phenotypic and molecular readouts
- Identify functional biomarkers of sensitivity or resistance
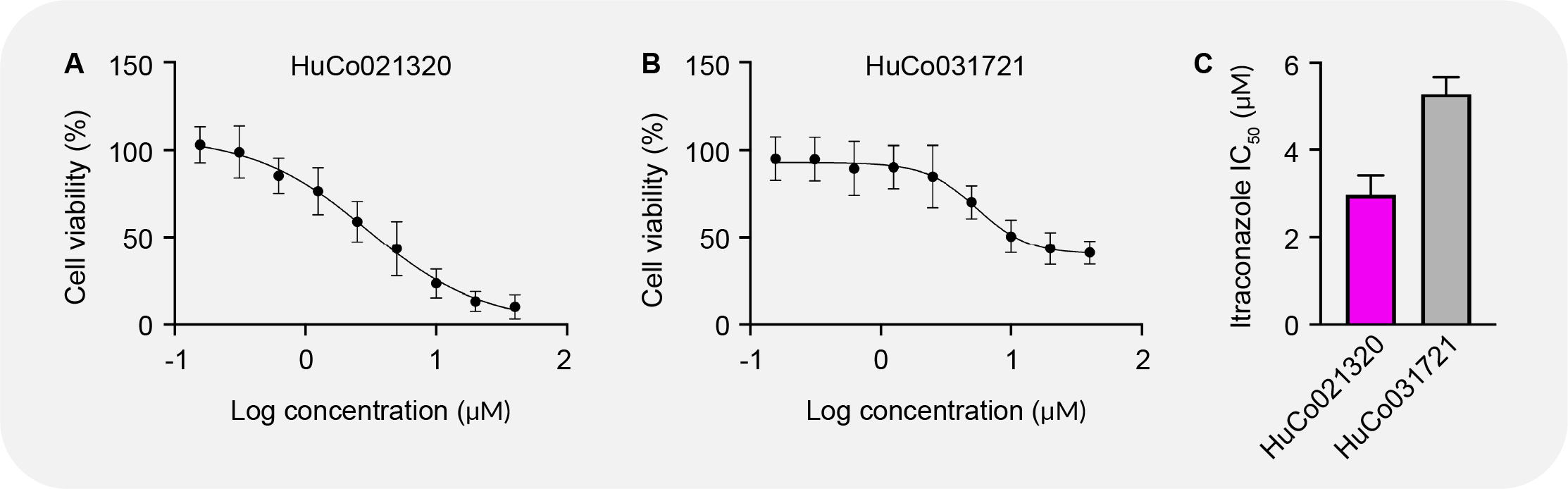
Image illustrates tumoroid response to targeted therapies. Itraconazole responses for (A) HuCo021320 and (B) HuCo031721. (C) Itraconazole IC50 values for HuCo021320 and HuCo031721 tumoroid lines.
Advancing translational pipelines
Bridge the gap between discovery and the clinic.
RASTRUM Allegro supports translational research teams, iPSC cores, and biopharma innovators by enabling the creation of consistent, biologically relevant 3D models that integrate seamlessly into downstream workflows. With support for long-term viability and standardized output, RASTRUM empowers cross-site collaboration, reproducibility, and preclinical progress.
- Generate standardized 3D models compatible with multiwell screening and multi-omics analysis
- Maintain long-term culture viability for chronic disease studies and therapeutic evaluation
- Enable reproducible results across teams, time points, and platforms
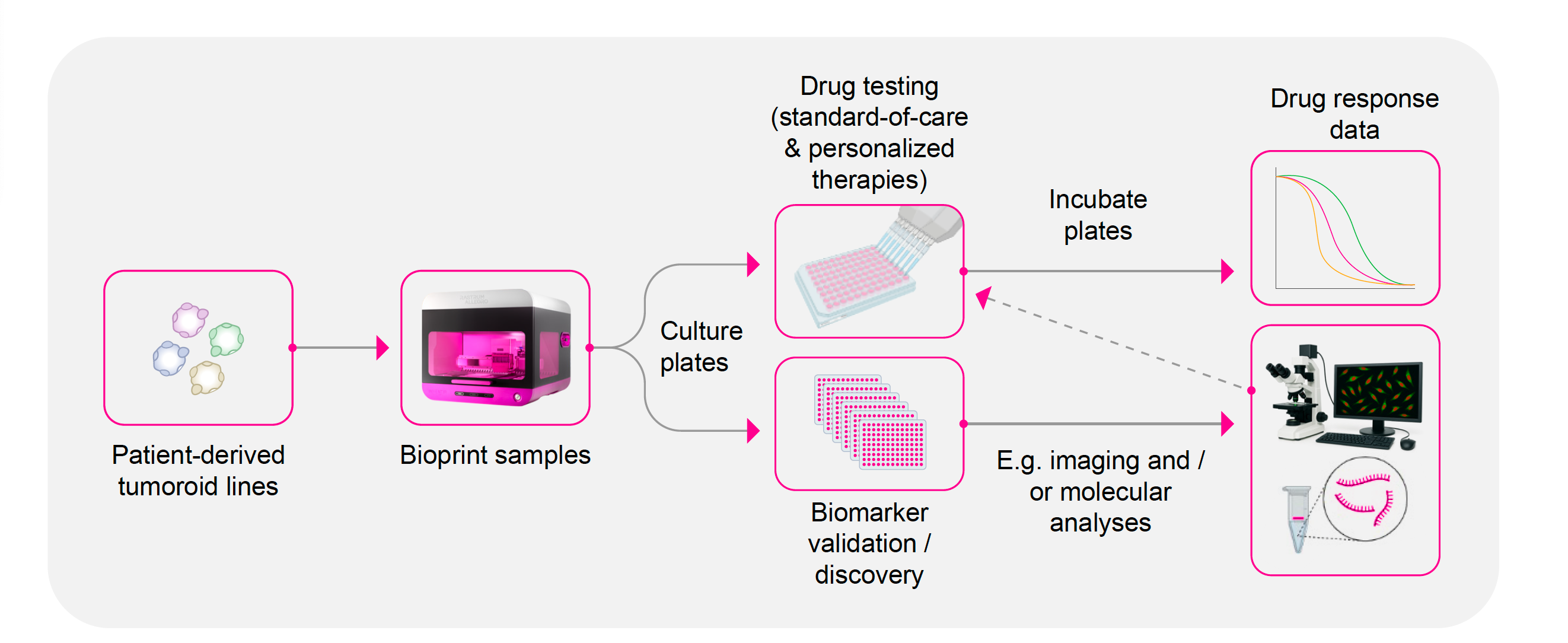
The infographic illustrates an example of how RASTRUM Allegro seamlessly integrates into translational pipelines in the precision medicine space for drug testing of standard of care and personalized therapies and biomarker validation and discovery work.
FAQs
Yes. RASTRUM was designed to support primary, patient-derived, and iPSC-based models, even when sample input is limited. Our matrix systems help preserve phenotype and promote long-term viability—making them ideal for translational and personalized medicine workflows.
Yes. RASTRUM’s ability to maintain molecular fidelity while supporting scalable screening makes it ideal for identifying functional biomarkers, stratifying patient responses, and validating treatment strategies across disease subtypes.
Absolutely. RASTRUM Matrices are tunable for stiffness, and biochemical cues—supporting disease-specific modeling across tissues such as brain, tumor microenvironments, and fibrotic niches.
Yes. RASTRUM outputs are assay-ready and compatible with high-content imaging, viability assays, RNA-seq, single-cell transcriptomics, and other multi-omics pipelines. The platform is designed to integrate seamlessly into downstream data analysis and discovery workflows.
RASTRUM Allegro delivers highly consistent models across wells, plates, and replicates. Our published data show low intra- and inter-plate coefficient of variation (CV), even in complex, patient-derived cultures.